Difference between revisions of "Lena Week 5"
(added screen shots) |
(→Reflections) |
||
| (25 intermediate revisions by one user not shown) | |||
| Line 1: | Line 1: | ||
==Electronic Journal for Uniprot exercise== | ==Electronic Journal for Uniprot exercise== | ||
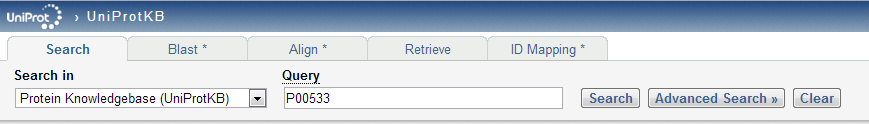
| − | *searched for primary accession number: P00533 | + | *'''searched for primary accession number: P00533''' |
| + | : | ||
[[Image:Lena1.PNG]] | [[Image:Lena1.PNG]] | ||
| − | *This is what the entry showed.. | + | : |
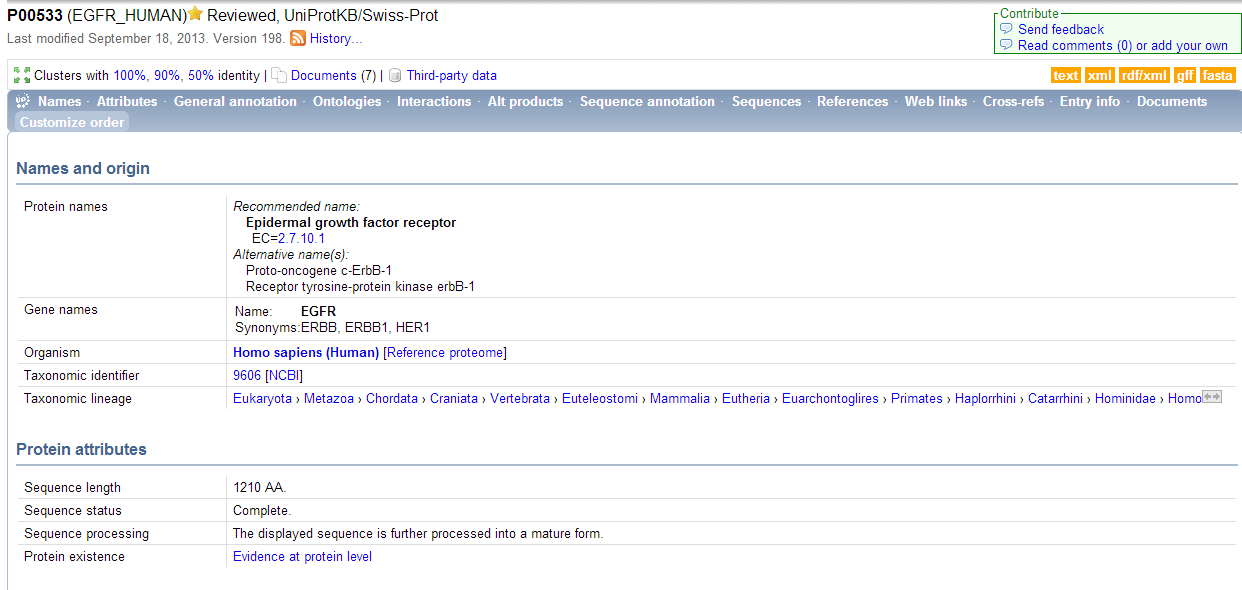
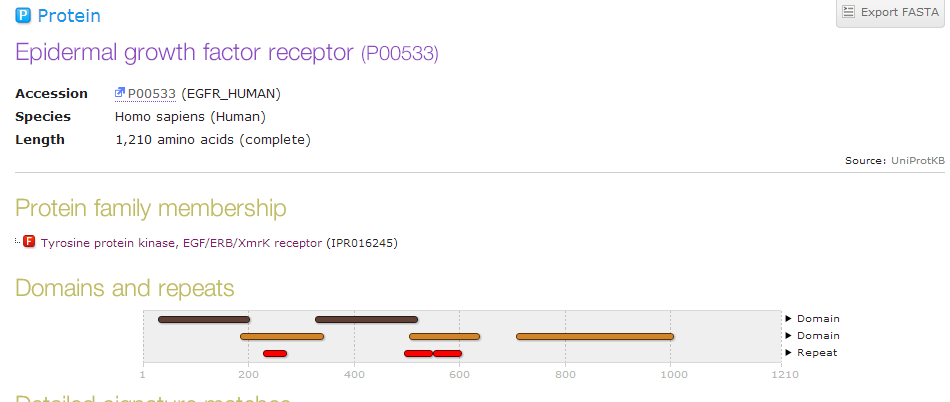
| + | *'''This is what the entry showed. It it the main page containing general information about EGFR.''' | ||
| + | : | ||
[[Image:Lena2.PNG]] | [[Image:Lena2.PNG]] | ||
| + | : | ||
| + | : | ||
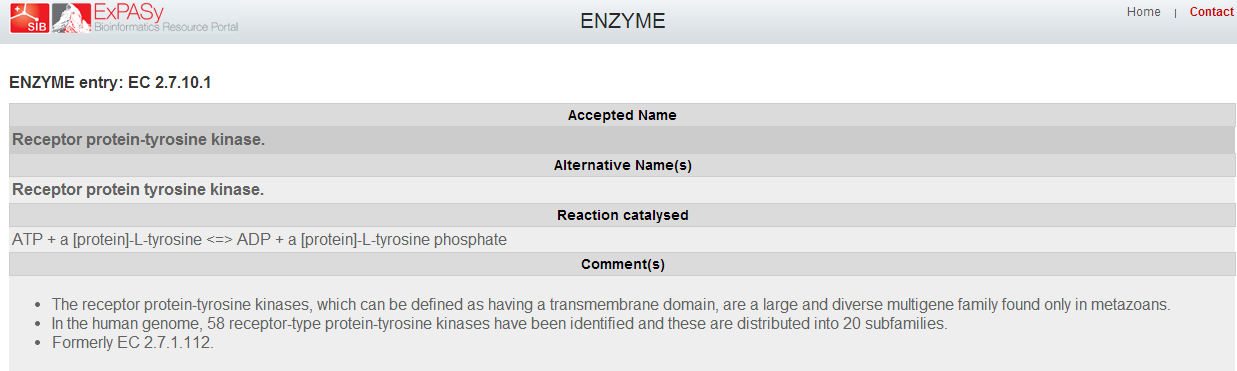
| + | *'''Clicked on EC 2.7.1.112 link, the link brought up this page. This page can help a person figure out a protein's enzymatic function.''' | ||
| + | : | ||
| + | : | ||
| + | :[[Image:Lena3.PNG]] | ||
| + | : | ||
| + | : | ||
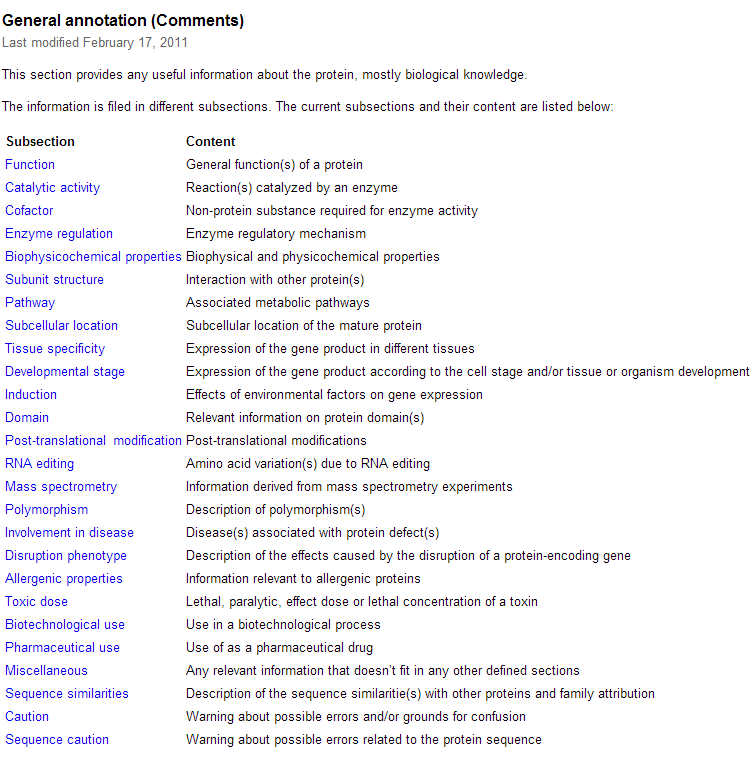
| + | *'''I navigated back to the original page, then clicked on the Comments heading to see what the comments where. I was directed to this page. The comments page hold useful information that just doesn't fit under any of the other headings.''' | ||
| + | : | ||
| + | : | ||
| + | :[[Image:Lena4.PNG]] | ||
| + | : | ||
| + | : | ||
| + | *'''Again, I went back to the main page and scrolled down to the cross reference section. I found InterPro and clicked on a link for Graphical View. The page showed a visual for the domains and repeats of the protein.''' | ||
| + | |||
| + | |||
| + | : | ||
| + | : | ||
| + | :[[Image:Lena5.PNG]] | ||
| + | : | ||
| + | : | ||
| + | : | ||
| + | : | ||
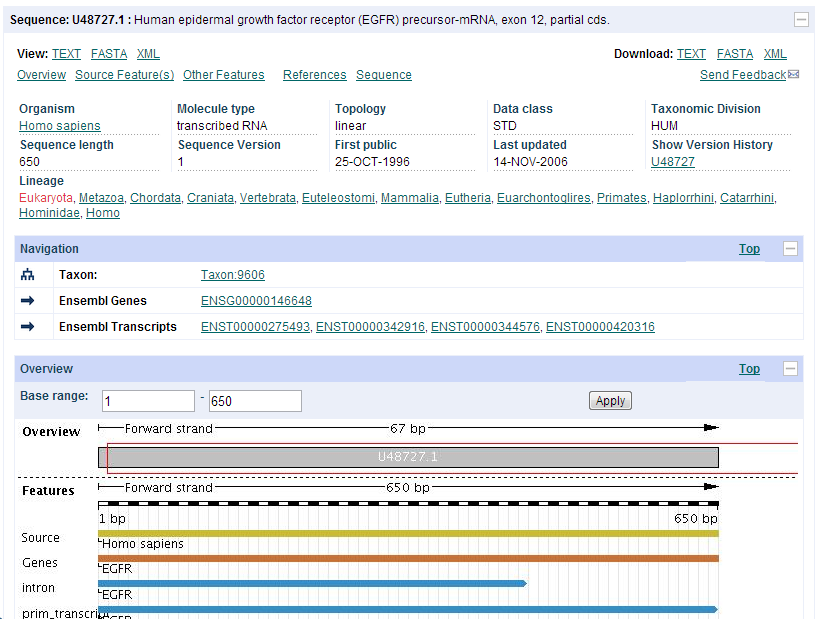
| + | *'''I also clicked on EMBL, this is the page that showed up. EMBL showed some basic information about the sequence I clicked on, and showed where repeats occured on the gene.''' | ||
| + | : | ||
| + | : | ||
| + | [[Image:Lena9.PNG]] | ||
| + | : | ||
| + | : | ||
| + | : | ||
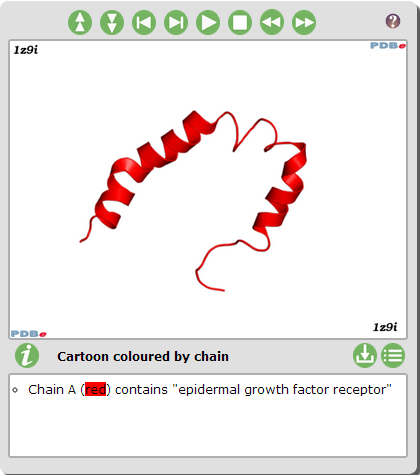
| + | *'''I looked at PDB which gave an animation of the protein. I liked this page because it showed you the different domains on the protein.''' | ||
| + | : | ||
| + | : | ||
| + | [[Image:Lena10.PNG]] | ||
| + | : | ||
| + | : | ||
| + | : | ||
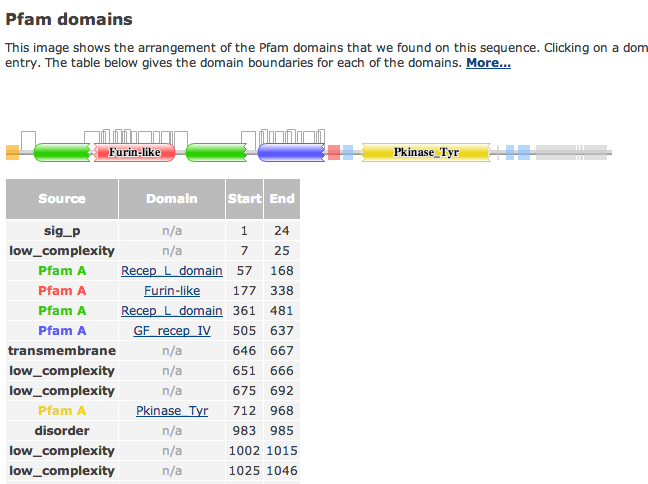
| + | *'''For the Pfam Link, I clicked on the graphical view and got this. The image shows the arrangement of Pfam domains on the P00533 sequence.''' | ||
| + | : | ||
| + | :[[Image:Pfam.PNG]] | ||
| + | : | ||
| + | : | ||
| + | *'''I tried out RefSeq, which gave a list of papers where the sequence was referenced. This could be useful in finding out more for a research project.''' | ||
| + | : | ||
| + | : | ||
| + | :[[Image:Lena12.PNG]] | ||
| + | |||
| + | |||
| + | : | ||
| + | : | ||
| + | *'''Lastly, under the Cross-refs, I tried out GeneID, which gave me this. It gave some general information and showed where the sequence was located genomically.''' | ||
| + | : | ||
| + | : | ||
| + | [[Image:GeneID.png]] | ||
| + | : | ||
| + | : | ||
| + | *'''Back on the main page, under the heading Ontologies, there were some keywords listed. I clicked on the keyword Tumor Supressor.''' | ||
| + | : | ||
| + | : | ||
| + | [[Image:Lena6.PNG]] | ||
| + | : | ||
| + | : | ||
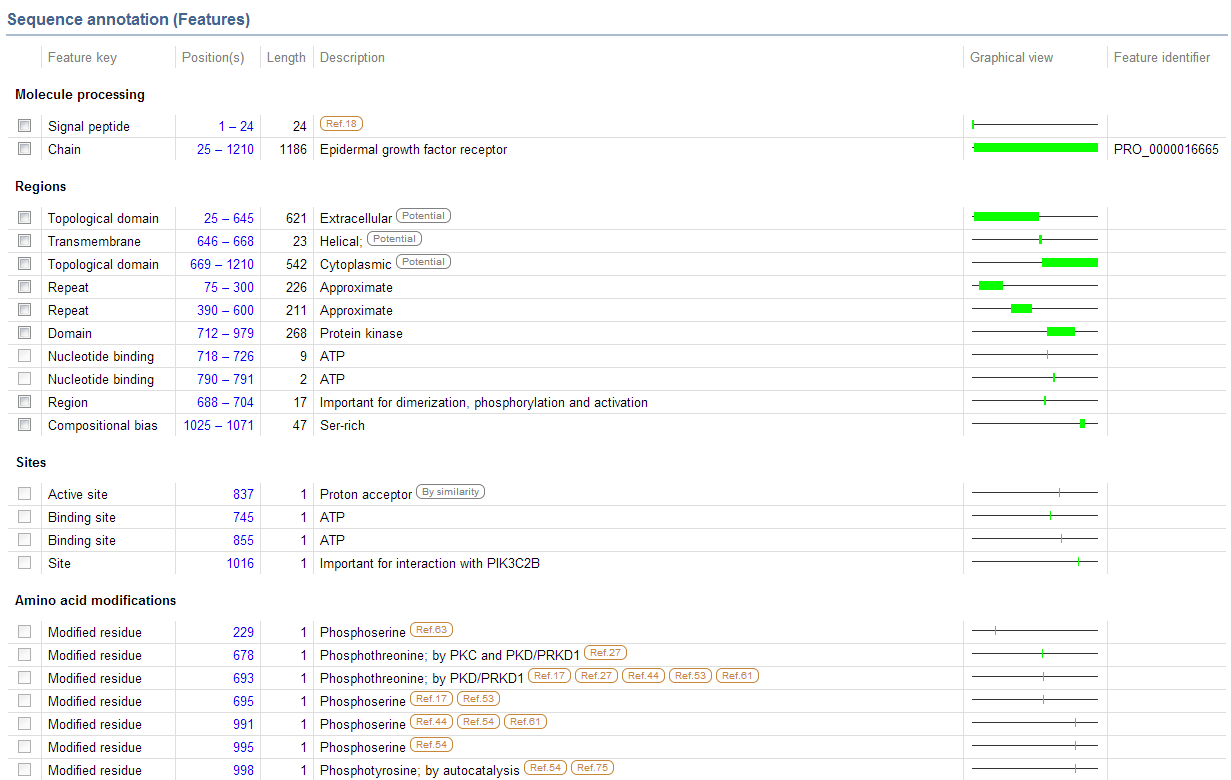
| + | *'''On the main page, under Sequence Annotation, this is what shows up''' | ||
| + | : | ||
| + | : | ||
| + | [[Image:Lena7.PNG]] | ||
| + | : | ||
| + | : | ||
| + | : | ||
| + | *'''I clicked on FASTA on the top right corner to get protein sequence in plain text. Here is the result''' | ||
| + | : | ||
| + | : | ||
| + | : | ||
| + | :>sp|P00533|EGFR_HUMAN Epidermal growth factor receptor OS=Homo sapiens GN=EGFR PE=1 SV=2 | ||
| + | MRPSGTAGAALLALLAALCPASRALEEKKVCQGTSNKLTQLGTFEDHFLSLQRMFNNCEV | ||
| + | VLGNLEITYVQRNYDLSFLKTIQEVAGYVLIALNTVERIPLENLQIIRGNMYYENSYALA | ||
| + | VLSNYDANKTGLKELPMRNLQEILHGAVRFSNNPALCNVESIQWRDIVSSDFLSNMSMDF | ||
| + | QNHLGSCQKCDPSCPNGSCWGAGEENCQKLTKIICAQQCSGRCRGKSPSDCCHNQCAAGC | ||
| + | TGPRESDCLVCRKFRDEATCKDTCPPLMLYNPTTYQMDVNPEGKYSFGATCVKKCPRNYV | ||
| + | VTDHGSCVRACGADSYEMEEDGVRKCKKCEGPCRKVCNGIGIGEFKDSLSINATNIKHFK | ||
| + | NCTSISGDLHILPVAFRGDSFTHTPPLDPQELDILKTVKEITGFLLIQAWPENRTDLHAF | ||
| + | ENLEIIRGRTKQHGQFSLAVVSLNITSLGLRSLKEISDGDVIISGNKNLCYANTINWKKL | ||
| + | FGTSGQKTKIISNRGENSCKATGQVCHALCSPEGCWGPEPRDCVSCRNVSRGRECVDKCN | ||
| + | LLEGEPREFVENSECIQCHPECLPQAMNITCTGRGPDNCIQCAHYIDGPHCVKTCPAGVM | ||
| + | GENNTLVWKYADAGHVCHLCHPNCTYGCTGPGLEGCPTNGPKIPSIATGMVGALLLLLVV | ||
| + | ALGIGLFMRRRHIVRKRTLRRLLQERELVEPLTPSGEAPNQALLRILKETEFKKIKVLGS | ||
| + | GAFGTVYKGLWIPEGEKVKIPVAIKELREATSPKANKEILDEAYVMASVDNPHVCRLLGI | ||
| + | CLTSTVQLITQLMPFGCLLDYVREHKDNIGSQYLLNWCVQIAKGMNYLEDRRLVHRDLAA | ||
| + | RNVLVKTPQHVKITDFGLAKLLGAEEKEYHAEGGKVPIKWMALESILHRIYTHQSDVWSY | ||
| + | GVTVWELMTFGSKPYDGIPASEISSILEKGERLPQPPICTIDVYMIMVKCWMIDADSRPK | ||
| + | FRELIIEFSKMARDPQRYLVIQGDERMHLPSPTDSNFYRALMDEEDMDDVVDADEYLIPQ | ||
| + | QGFFSSPSTSRTPLLSSLSATSNNSTVACIDRNGLQSCPIKEDSFLQRYSSDPTGALTED | ||
| + | SIDDTFLPVPEYINQSVPKRPAGSVQNPVYHNQPLNPAPSRDPHYQDPHSTAVGNPEYLN | ||
| + | TVQPTCVNSTFDSPAHWAQKGSHQISLDNPDYQQDFFPKEAKPNGIFKGSTAENAEYLRV | ||
| + | APQSSEFIGA | ||
| + | : | ||
| + | ==About the EGFR-HUMAN Protein== | ||
| + | :The EGTR_HUMAN is a protein know as "epidermal growth factor receptor." The Epidermal growth factor and its receptor were found at Vanderbilt University by researcher Stanely Cohen. It is a member of the kinase protein superfamily and the subfamily of ErbB receptor. The protein in located on the plasma membrane and binds to ligands, including an epidermal growth factor and a transforming growth factor. Mutations affecting this protein could lead to cancer, particularly in the lungs. | ||
| + | |||
| + | ==Reflections== | ||
| + | #The purpose of the exercise was to get familiar with the layout and features of Uniprot, especially because we are going to be working with Uniprot a lot more this semester. | ||
| + | #I learned a bit about which cross-ref links I liked, and how to go about accessing information about a specific protein. | ||
| + | #I think I understood the exercise, but the information being present by some of the cross-ref pages seems really in depth and over my head. I feel like I don't yet know how to interpret all the information I am receiving. | ||
| + | |||
| + | : | ||
| + | [[User:Lena|Lena]] ([[User talk:Lena|talk]]) 17:25, 26 September 2013 (PDT) | ||
| + | :[[Category:Journal Entry]] | ||
Latest revision as of 00:25, 27 September 2013
[edit] Electronic Journal for Uniprot exercise
- searched for primary accession number: P00533
- This is what the entry showed. It it the main page containing general information about EGFR.
- Clicked on EC 2.7.1.112 link, the link brought up this page. This page can help a person figure out a protein's enzymatic function.
- I navigated back to the original page, then clicked on the Comments heading to see what the comments where. I was directed to this page. The comments page hold useful information that just doesn't fit under any of the other headings.
- Again, I went back to the main page and scrolled down to the cross reference section. I found InterPro and clicked on a link for Graphical View. The page showed a visual for the domains and repeats of the protein.
- I also clicked on EMBL, this is the page that showed up. EMBL showed some basic information about the sequence I clicked on, and showed where repeats occured on the gene.
- I looked at PDB which gave an animation of the protein. I liked this page because it showed you the different domains on the protein.
- For the Pfam Link, I clicked on the graphical view and got this. The image shows the arrangement of Pfam domains on the P00533 sequence.
- I tried out RefSeq, which gave a list of papers where the sequence was referenced. This could be useful in finding out more for a research project.
- Lastly, under the Cross-refs, I tried out GeneID, which gave me this. It gave some general information and showed where the sequence was located genomically.
- Back on the main page, under the heading Ontologies, there were some keywords listed. I clicked on the keyword Tumor Supressor.
- On the main page, under Sequence Annotation, this is what shows up
- I clicked on FASTA on the top right corner to get protein sequence in plain text. Here is the result
- >sp|P00533|EGFR_HUMAN Epidermal growth factor receptor OS=Homo sapiens GN=EGFR PE=1 SV=2
MRPSGTAGAALLALLAALCPASRALEEKKVCQGTSNKLTQLGTFEDHFLSLQRMFNNCEV VLGNLEITYVQRNYDLSFLKTIQEVAGYVLIALNTVERIPLENLQIIRGNMYYENSYALA VLSNYDANKTGLKELPMRNLQEILHGAVRFSNNPALCNVESIQWRDIVSSDFLSNMSMDF QNHLGSCQKCDPSCPNGSCWGAGEENCQKLTKIICAQQCSGRCRGKSPSDCCHNQCAAGC TGPRESDCLVCRKFRDEATCKDTCPPLMLYNPTTYQMDVNPEGKYSFGATCVKKCPRNYV VTDHGSCVRACGADSYEMEEDGVRKCKKCEGPCRKVCNGIGIGEFKDSLSINATNIKHFK NCTSISGDLHILPVAFRGDSFTHTPPLDPQELDILKTVKEITGFLLIQAWPENRTDLHAF ENLEIIRGRTKQHGQFSLAVVSLNITSLGLRSLKEISDGDVIISGNKNLCYANTINWKKL FGTSGQKTKIISNRGENSCKATGQVCHALCSPEGCWGPEPRDCVSCRNVSRGRECVDKCN LLEGEPREFVENSECIQCHPECLPQAMNITCTGRGPDNCIQCAHYIDGPHCVKTCPAGVM GENNTLVWKYADAGHVCHLCHPNCTYGCTGPGLEGCPTNGPKIPSIATGMVGALLLLLVV ALGIGLFMRRRHIVRKRTLRRLLQERELVEPLTPSGEAPNQALLRILKETEFKKIKVLGS GAFGTVYKGLWIPEGEKVKIPVAIKELREATSPKANKEILDEAYVMASVDNPHVCRLLGI CLTSTVQLITQLMPFGCLLDYVREHKDNIGSQYLLNWCVQIAKGMNYLEDRRLVHRDLAA RNVLVKTPQHVKITDFGLAKLLGAEEKEYHAEGGKVPIKWMALESILHRIYTHQSDVWSY GVTVWELMTFGSKPYDGIPASEISSILEKGERLPQPPICTIDVYMIMVKCWMIDADSRPK FRELIIEFSKMARDPQRYLVIQGDERMHLPSPTDSNFYRALMDEEDMDDVVDADEYLIPQ QGFFSSPSTSRTPLLSSLSATSNNSTVACIDRNGLQSCPIKEDSFLQRYSSDPTGALTED SIDDTFLPVPEYINQSVPKRPAGSVQNPVYHNQPLNPAPSRDPHYQDPHSTAVGNPEYLN TVQPTCVNSTFDSPAHWAQKGSHQISLDNPDYQQDFFPKEAKPNGIFKGSTAENAEYLRV APQSSEFIGA
[edit] About the EGFR-HUMAN Protein
- The EGTR_HUMAN is a protein know as "epidermal growth factor receptor." The Epidermal growth factor and its receptor were found at Vanderbilt University by researcher Stanely Cohen. It is a member of the kinase protein superfamily and the subfamily of ErbB receptor. The protein in located on the plasma membrane and binds to ligands, including an epidermal growth factor and a transforming growth factor. Mutations affecting this protein could lead to cancer, particularly in the lungs.
[edit] Reflections
- The purpose of the exercise was to get familiar with the layout and features of Uniprot, especially because we are going to be working with Uniprot a lot more this semester.
- I learned a bit about which cross-ref links I liked, and how to go about accessing information about a specific protein.
- I think I understood the exercise, but the information being present by some of the cross-ref pages seems really in depth and over my head. I feel like I don't yet know how to interpret all the information I am receiving.
Lena (talk) 17:25, 26 September 2013 (PDT)