Difference between revisions of "Gleis Week 12"
From LMU BioDB 2013
| (13 intermediate revisions by one user not shown) | |||
| Line 1: | Line 1: | ||
| + | ==Lab Journal== | ||
| + | *Checked out code from sourceforge @ https://svn.code.sf.net/p/xmlpipedb/code | ||
| + | *Followed directions to configure new project wizard | ||
| + | *Entered name and clicked finish | ||
| + | *Add JDK work environment | ||
| + | :*Window>preferences>Java | ||
| + | *Follow direction to build path and use as source folder | ||
| + | *Begin adding species profile | ||
| + | *src folder> edu.lmu.xmlpipedb.gmbuilder.databasetoolkit.profiles | ||
| + | *right click and New > Class | ||
| + | *enter the following: | ||
| + | Name: LeishmaniaMajorUniProtSpeciesProfile (no spaces, capitalizing the first letters of each word) | ||
| + | Superclass: edu.lmu.xmlpipedb.gmbuilder.databasetoolkit.profiles.UniProtSpeciesProfile (you can also click on Browse... to navigate to this if you don’t feel like typing) | ||
| + | *clicked finish | ||
| + | *open new java file | ||
| + | *add constructor from [[Coder]] | ||
| + | :*edit for Leishmania major | ||
| + | *Entered code for OrderedLocusNames | ||
| + | :*[[Coder]] | ||
| + | |||
| + | |||
Eclipse>workbench>+canister>follow coder instructions>window> | Eclipse>workbench>+canister>follow coder instructions>window> | ||
| + | |||
| + | |||
| + | notes: | ||
| + | use antbuild in eclipse | ||
| + | use firstobject xml | ||
| + | edit>find>F3 | ||
| + | |||
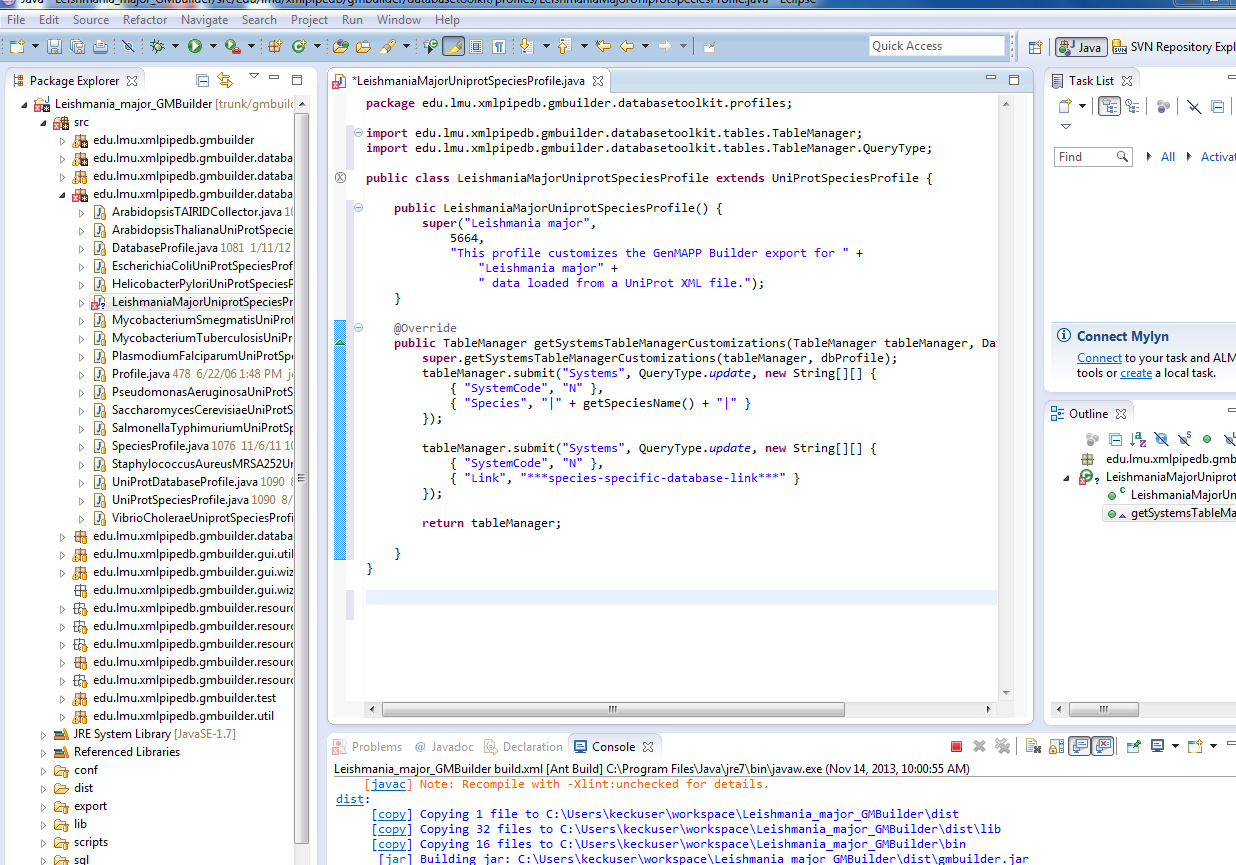
| + | customize | ||
| + | src>right click datbase profiles>follow coder page | ||
| + | |||
| + | :[[Image:Eclipse customs Gleis 14112013.PNG]] | ||
| + | |||
| + | Identify MOD- species pattern link | ||
| + | |||
| + | Run export and check for database update | ||
| + | |||
| + | [[User:Gleis|Gleis]] ([[User talk:Gleis|talk]]) 11:54, 14 November 2013 (PST) | ||
| + | |||
| + | [[Leishmania major|Off The Leish]] | ||
| + | |||
| + | [[Category:Journal Entry]] [[Category:Leishmania major]] | ||
Latest revision as of 19:58, 14 November 2013
[edit] Lab Journal
- Checked out code from sourceforge @ https://svn.code.sf.net/p/xmlpipedb/code
- Followed directions to configure new project wizard
- Entered name and clicked finish
- Add JDK work environment
- Window>preferences>Java
- Follow direction to build path and use as source folder
- Begin adding species profile
- src folder> edu.lmu.xmlpipedb.gmbuilder.databasetoolkit.profiles
- right click and New > Class
- enter the following:
Name: LeishmaniaMajorUniProtSpeciesProfile (no spaces, capitalizing the first letters of each word) Superclass: edu.lmu.xmlpipedb.gmbuilder.databasetoolkit.profiles.UniProtSpeciesProfile (you can also click on Browse... to navigate to this if you don’t feel like typing)
- clicked finish
- open new java file
- add constructor from Coder
- edit for Leishmania major
- Entered code for OrderedLocusNames
Eclipse>workbench>+canister>follow coder instructions>window>
notes:
use antbuild in eclipse
use firstobject xml
edit>find>F3
customize src>right click datbase profiles>follow coder page
Identify MOD- species pattern link
Run export and check for database update