Difference between revisions of "Gene Database Project"
Kdahlquist (Talk | contribs) (created category for group projects) |
Kdahlquist (Talk | contribs) (→Group: added gene database testing report) |
||
| Line 12: | Line 12: | ||
** Include Gene Database Schema diagram in ReadMe | ** Include Gene Database Schema diagram in ReadMe | ||
*** Sample schema in Adobe Illustrator format: [[Media:Vibrio_schema_20101022.zip | Vibrio_schema_20101022.zip]] | *** Sample schema in Adobe Illustrator format: [[Media:Vibrio_schema_20101022.zip | Vibrio_schema_20101022.zip]] | ||
| + | * Gene Database Testing Report for final submitted Gene Database (print from wiki to ''.pdf'' file) | ||
* Processed and analyzed DNA microarray dataset (''.xls'') | * Processed and analyzed DNA microarray dataset (''.xls'') | ||
* GenMAPP Expression Dataset file (''.gex'') | * GenMAPP Expression Dataset file (''.gex'') | ||
| Line 17: | Line 18: | ||
* Sample MAPP file of a relevant biological pathway for your species (''.mapp'') | * Sample MAPP file of a relevant biological pathway for your species (''.mapp'') | ||
* [[Gene Database Project Report Guidelines#Group Report | Group Report]] describing the creation of the Gene Database and the biological analysis of the data (''.doc'' or ''.pdf'') | * [[Gene Database Project Report Guidelines#Group Report | Group Report]] describing the creation of the Gene Database and the biological analysis of the data (''.doc'' or ''.pdf'') | ||
| − | * PowerPoint presentation (''.ppt'', given | + | * PowerPoint presentation (''.ppt'', given on Thursday, December 12) |
| − | + | ||
=== Individual === | === Individual === | ||
Revision as of 19:22, 18 October 2013
Contents |
Deliverables
- You will give a final group PowerPoint presentation in class on Thursday, December 12, 8:00am.
- Final due date for all other deliverables is no later than Friday, December 13, 4:30pm (according to official final exam schedule for fall 2013)
- The deliverables should be uploaded and organized onto one group wiki page, or alternately, delivered on CD to either Dr. Dahlquist or Dr. Dionisio.
- Each member of the group will be assigned the same grade for the group project.
Group
- GenMAPP Gene Database for assigned species (.gdb)
- ReadMe file to accompany the Gene Database (.pdf)
- Sample ReadMe in Word format: ReadMe_Vc-Std_External_20101022.zip
- Include Gene Database Schema diagram in ReadMe
- Sample schema in Adobe Illustrator format: Vibrio_schema_20101022.zip
- Gene Database Testing Report for final submitted Gene Database (print from wiki to .pdf file)
- Processed and analyzed DNA microarray dataset (.xls)
- GenMAPP Expression Dataset file (.gex)
- Filtered MAPPFinder Results (.xls)
- Sample MAPP file of a relevant biological pathway for your species (.mapp)
- Group Report describing the creation of the Gene Database and the biological analysis of the data (.doc or .pdf)
- PowerPoint presentation (.ppt, given on Thursday, December 12)
Individual
- Assessment and reflection on process
- Statement of work
- Assessment of the work done
- What was learned
Team Journal Entries
- Each team will write a combined journal entry for each week with contributions from all members.
- Week 11 (midnight 11/8)
- Week 12 (midnight 11/15)
- Week 13 (midnight 11/22)
- Week 15 (midnight 12/6)
Groups
The project groups are:
- To be determined.
Roles (Guilds)
As the project moves forward, we will use class time for team meetings. In addition, we will also have guild meetings where students sharing the same role can work together on common issues. Each student has been assigned a primary role in the project by the instructors. The student who was elected the team leader also has a secondary role as the project manager.
Project Manager (PM)
The project manager makes sure that individuals are fulfilling their roles and performing the tasks on time.
GenMAPP User (GU)
The GenMAPP user runs GenMAPP with the microarray dataset (Expression Dataset Manager, MAPPFinder, GenMAPP MAPP).
Quality Assurance (QA)
The Quality Assurance team member is the resident expert on species ID systems and formats. He or she should be proficient with XMLPipeDB Match, SQL queries in PostgreSQL, and Microsoft Access to navigate through the data and find missing IDs, discrepancies, sanity checks, etc.
Coder (C)
The coder is the resident expert on the technology being used — assorted software, file management, some troubleshooting, possibly some programming. He or she coordinates with Drs. Dahlquist and Dionisio in extending GenMAPP Builder code and making new versions
Project Milestones
The project milestones have been moved to the individual guild pages.
Overall Flow
A successful project will have the following steps from end to end. Depending on the data and/or other issues that are encountered, one or more of these steps may go through iterations, repetitions, or refinements.
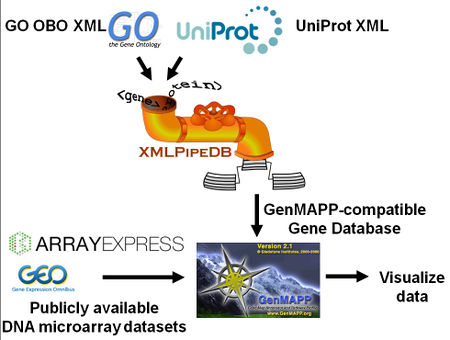
- Following the steps shown in the Running GenMAPP Builder instructions, download the UniProt XML proteome set and GOA (GO association) files from Integr8 for your species.
- Make sure that you are choosing the correct strain/subspecies as the microarray data you have.
- Note the date of download and the version of the file from the status page.
- Download GO terms from Gene Ontology.
- You will need the OBO-XML format, the "obo-xml.gz" file.
- Note the version of your file (date and time of last update are shown at the top of the page).
- Create the GenMAPP Builder tables in PostgreSQL.
- Load files into PostgreSQL database via GenMAPP Builder.
- Export into a GenMAPP Gene Database.
- Inspect/vet/validate Gene Database.
- Create a Gene Database Testing Report each time a new export is run.
- Compare to the model organism database for your species
- Find an original journal article describing a DNA microarray experiment performed on your species.
- Download the microarray data for this article.
- Consult with Dr. Dahlquist about the proper steps to take to process the data (normalization, statistical analysis) and perform the analysis
- Run GenMAPP using the Gene Database.
- Microarray data (import using Expression Dataset Manager)
- MAPPFinder
- Place genes on MAPP and draw pathway
Guild Milestones
These are links to the respective milestone lists in the guild pages.