Difference between revisions of "Alina's Assessment and Reflection"
From LMU BioDB 2013
(→Statement of Work: visual help info) |
(→Statement of Work) |
||
| Line 4: | Line 4: | ||
===Updating of Gene Database Testing Reports=== | ===Updating of Gene Database Testing Reports=== | ||
| − | *Was responsible for running counts (XMLPipeDB Match, Tally Engine, SQL) | + | *Was responsible for running counts (XMLPipeDB Match, Tally Engine, SQL)for all export testing reports |
*Performed visual inspections within gdb files | *Performed visual inspections within gdb files | ||
**Systems table | **Systems table | ||
| Line 10: | Line 10: | ||
**RefSeq table | **RefSeq table | ||
**OriginalRowCounts | **OriginalRowCounts | ||
| − | * | + | *Used snipping tool to provide screenshots for most results in testing |
*(Did not update GenMAPP aspects, left to Kevin) | *(Did not update GenMAPP aspects, left to Kevin) | ||
*Compared found Gene ID formats with corresponding online resources (UniProt, RefSeq) | *Compared found Gene ID formats with corresponding online resources (UniProt, RefSeq) | ||
| Line 26: | Line 26: | ||
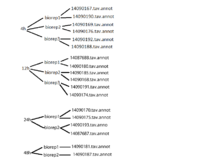
*Used Paint program to create diagram | *Used Paint program to create diagram | ||
[[Image:TATK tree.png|200px]] | [[Image:TATK tree.png|200px]] | ||
| + | |||
* Provide references or links to artifacts of your work, such as: | * Provide references or links to artifacts of your work, such as: | ||
Revision as of 22:19, 13 December 2013
Contents |
Statement of Work
- Describe exactly what you did on the project.
Updating of Gene Database Testing Reports
- Was responsible for running counts (XMLPipeDB Match, Tally Engine, SQL)for all export testing reports
- Performed visual inspections within gdb files
- Systems table
- UniProt table
- RefSeq table
- OriginalRowCounts
- Used snipping tool to provide screenshots for most results in testing
- (Did not update GenMAPP aspects, left to Kevin)
- Compared found Gene ID formats with corresponding online resources (UniProt, RefSeq)
- Investigated Ensembl (MOD) to compare gene totals
- Export Reports contributed to:
XML/Exception File Comparison
- Used XMLPipeDB Match in order to search Uniprot XML file for species IDs with format SP_[0-9][0-9][0-9][0-9] and got 2126 results.
- This was saved as a text file in order to make compatible for Excel. This file and exceptions file were put in adjacent columns and Excel match function was used. All results found to be #N/A.
- Excel File: ID Comparison
Visual Representation of Data Information
- Helped Kevin with visual representating replicate information from microarray paper
- Used Paint program to create diagram
- Provide references or links to artifacts of your work, such as:
- Wiki pages
- Other files or documents
- Code or scripts
Assessment of Project
- Give an objective assessment of the success of your project workflow and teamwork.
- What worked and what didn't work?
- What would you do differently if you could do it all over again?
- Evaluate the Gene Database Project and Group Report in the following areas:
- Content: What is the quality of the work?
- Organization: Comment on the organization of the project and of your group's wiki pages.
- Completeness: Did your team achieve all of the project objectives? Why or why not?
Reflection on the Process
- What did you learn?
- With your head (biological or computer science principles)
- With your heart (personal qualities and teamwork qualities that make things work or not work)?
- With your hands (technical skills)?
- What lesson will you take away from this project that you will still use a year from now?