HDelgadi Week 5
UniProt Exercise
In order to search for the ID P00533, which is known as the primary accession number and looks is used to search for the corresponding protein in the database, I used the following URL: http://www.uniprot.org/. I typed in the given ID number, P00533, under the Query subheading at the top near the "Search" button to its right. The search button was clicked on and the following information about this ID number which stands for the protein of interest was shown,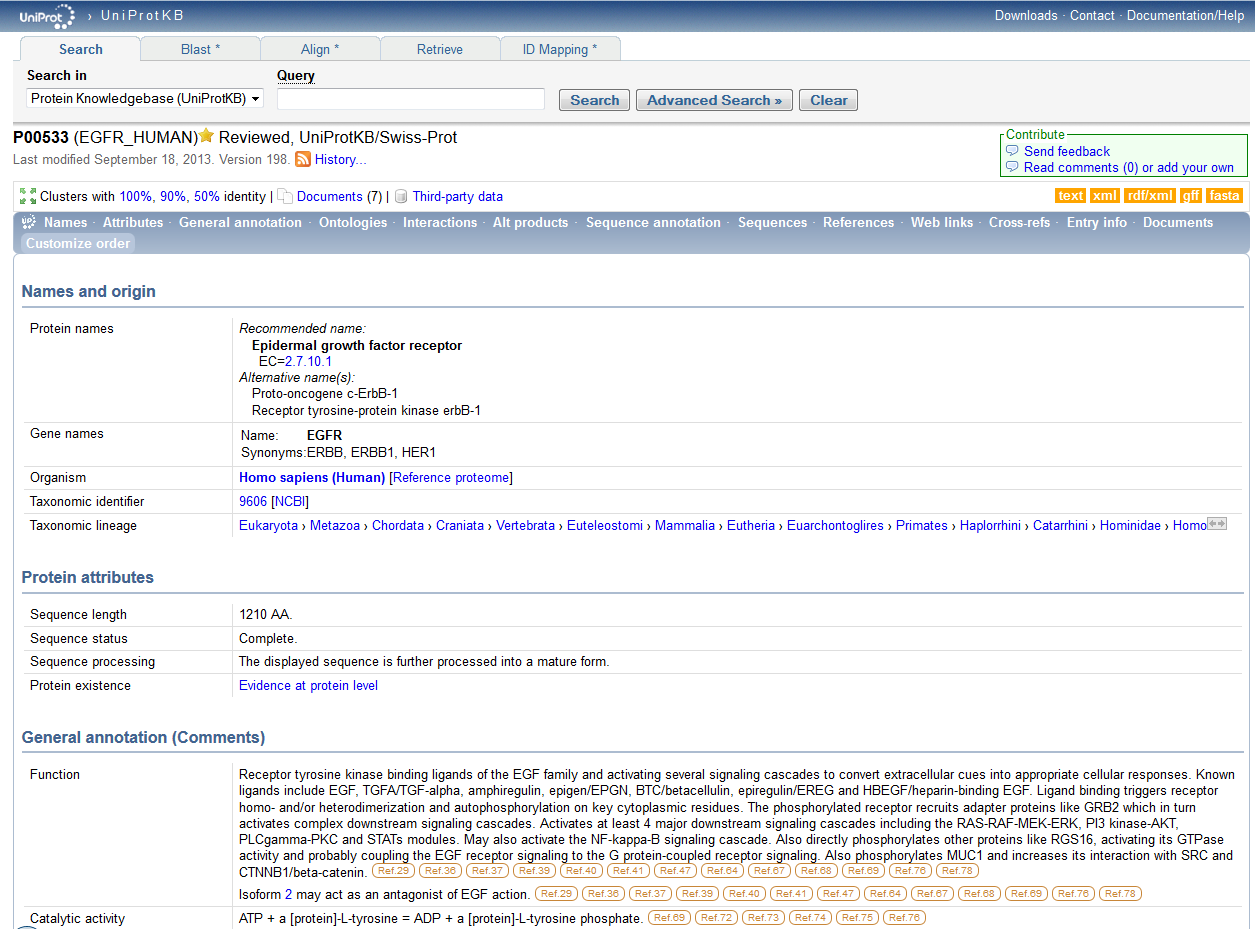
In order to get to the "ENZYME entry: EC 2.7.10.1" page which is referred to as the NiceZyme view of the enzyme in the reading, I had to click on EC=2.7.10.1 on the right of "Protein names" and under Recommended name. The subsequent page is the one as shown, 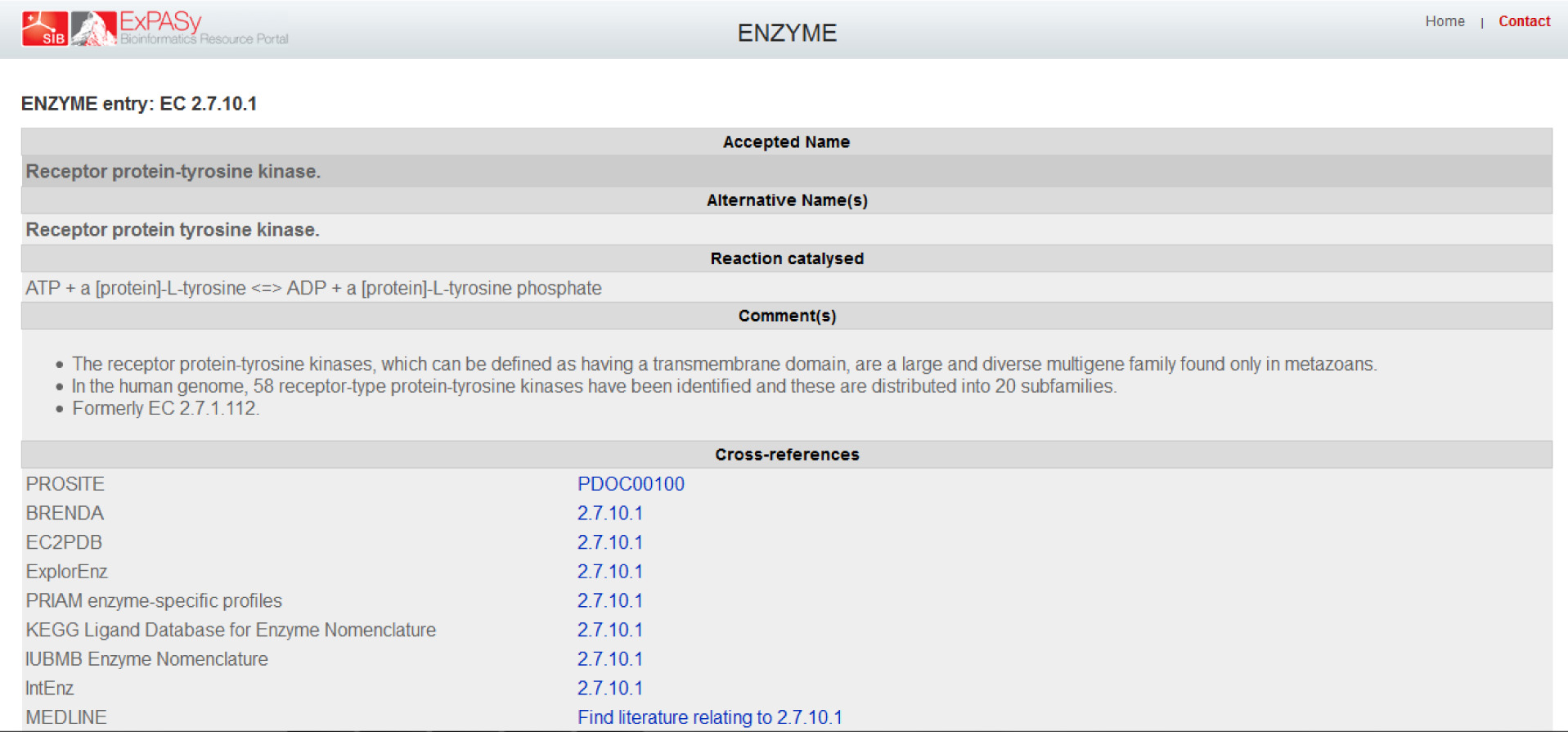
Furthermore, if I click on 9606 that is positioned to the right of "Taxonomic identifier", I can assess the taxonomy of where the protein is found, in this case in Humans. The page after clicking the Taxonomic identifier should look like this, Expasy 2.jpg.
Write a one-paragraph summary of what you have learned about the human EGFR protein from this exercise. Reflect and answer the following questions on your individual journal page:
- What was the purpose of this exercise?
- What did I learn from this exercise?
- What did I not understand (yet) about this exercise?