Lena Project Notebook
From LMU BioDB 2013
Contents |
Week 12
Gabriel and I performed an import/export cycle on 11/14/2013.
Export Information
- Uniprot: 7.12 minutes
- Version: UniProt release 2013_10 - October 16, 2013
- File:UniprotXML Leishmania 05112013 Gabe Lena.xml
- GO OBO: 6.32 minutes
- Version: Monday, November 04, 2013, 2:03:38 AM
- File:Leishmania 05112013 Gabe Lena.obo-xml.gz
- GOA: 4.54 minutes
- Version: 14 November, 2013
- 12-Nov-2013 11:47 3.0M
- http://ftp.ebi.ac.uk/pub/databases/GO/goa/proteomes/21780.L_major.goa
- File:LeishmaniaGOA 19112013 Lena Gabe.goa
- Name of .gdb file
- Leishmania_05112013_Lena_Gabe.gdb
- File:Leishmania 05112013 Lena Gabe.gdb
Tally Engine
Using XMLPipeDB match to Validate the XML Results from the TallyEngine
Original Row Counts Comparison
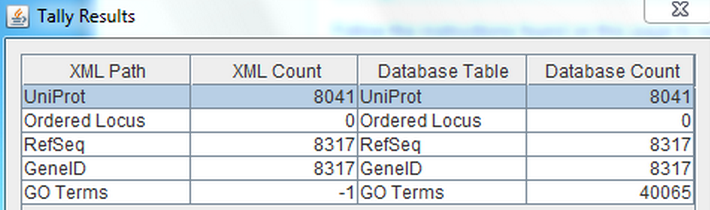
- Uniprot has 8041 which is the same the tallycount.
- There were 0 ordered locus, which is the same as the tallycount.
- There were 8315 hits for RefSeq, which is 2 fewer than was seen in the tallycount.
- There were 8315 hits for GeneID, which is 2 fewer than was seen in the tallycount.
Note:Leishmania major does not have "ordered locus names," instead they are tagged as "ORF."
File Management
- To keep the names of our files consistent, we agreed to name files with 1.) an identifier of what the file contains (such as "Uniprot") 2.) Leishmania 3.) the date in format MMDDYYYY 4.) names of Team members.
- An example of our file titles is: UniprotXML Leishmania 05112013 Gabe Lena.xml
- All files are stored on the main page for Leishmania major for ease of access. On the computer, the files are stored in the Downloads file.
System IDs
- OrderedLocusPattern: LmjF##.#### or LmjF_##_#### or LmjF.##.####
- Taxon ID: 5664
Week 13
- I updates the export files. The files now have the proper identification information to be found again. The GOA file had to re-downloaded to a more updated version and now has to be imported.
- The reason why no Ordered Locus Names turned up on the tally engine is that Leishmania major's Ordered Locus Names are tagged with ORF instead. Gabe re-coded to account for this.
- Built new database called Leishmania_major_18112013 and ran a new import/export cycle with the updated files.
- My target for this week was to get to know System IDs, and characterize regular expression patterns to detect the IDs. I found ID pattern to be: LmjF##.#### or LmjF_##_#### or LmjF.##.####
- A customized database was built for Leishmania major File:Leishmania major 19112013 Dist.zip
Export Information
- Uniprot: 7.42 minutes
- Version: UniProt release 2013_10 - October 16, 2013
- File:UniprotXML Leishmania 05112013 Gabe Lena.xml
- GO OBO: 5.96
- Gene Ontology Processing: 4.48 minutes
- Version: Monday, November 04, 2013, 2:03:38 AM
- File:Leishmania 05112013 Gabe Lena.obo-xml.gz
- NOTE:When we went to import this file we got an error message. The file that should be used was called Leishmania_05112013_Gabe_Lena.obo-xml. This file type is not uploadable to the wiki, and so only the zipped version is available. Just remember to use unzipped version for uploads.
- GOA: 0.04
- Version: 14 November, 2013
- 12-Nov-2013 11:47 3.0M
- http://ftp.ebi.ac.uk/pub/databases/GO/goa/proteomes/21780.L_major.goa
- File:LeishmaniaGOA 19112013 Lena Gabe.goa
- Name of .gdb file: LeishmaniaGDB_19112013_LenaLGabe/gdb
- Leishmania_05112013_Lena_Gabe.gdb
- File:Leishmania 05112013 Lena Gabe.gdb
11/21/13: The new database was labeled as generic when we tried to open it before. Today we tried to open a OrderedLocusName in GenMAPP but the gene was not found. The database has to be re-coded and re-exported. I edited the database in eclipse. When we exported to GenMAPP it finally recognized the database. We saved as Media:LeishmaniaGDB 21112013 Lena Gabe.gdb.
Week 14
- Configured Leishmania_major-18112013 in GenMAPP Builder, and ran Tally engine.
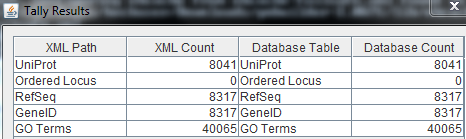
Tally Engine
- Finally, the numbers are matching, but the Ordered Locus names are still missing. This may be because Ordered Locus Names are called ORFs for Leishmania major.
XMLpipedb Match
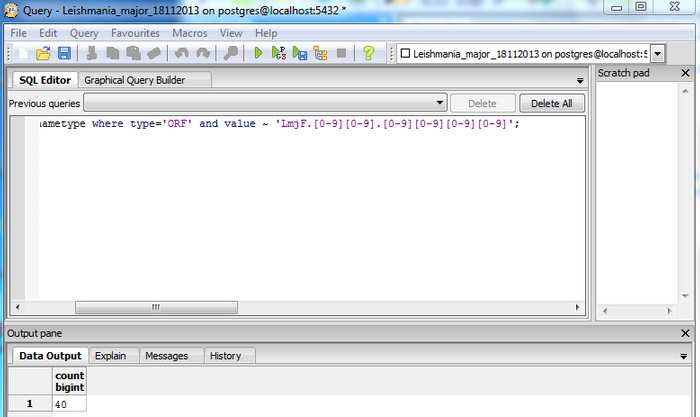
- We XML pipedb Match query to see if we could find the missing terms. "ORF" yielded 40 results; "ordered locus name" yielded 0 results.
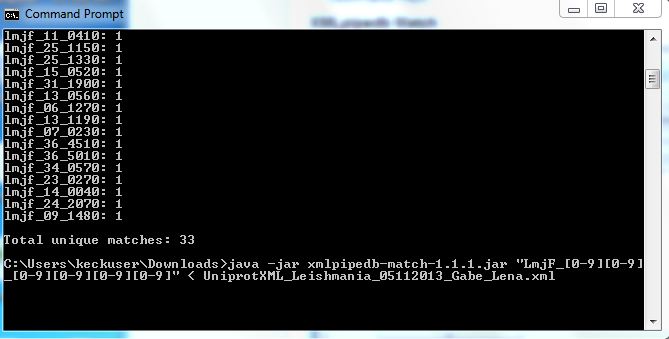
- Used Match in the command line. Found 33 matched to lmjf_##_####. Now we have to figure how to get the computer to get either an underscore or period in the name.
Week 15
GenMAPP Expression Dataset Manager
- Database: LeishmaniaGDB_26112013_Lena_Gabe.gdb
- File: LeishmaniaCompiledStatAnalysis(A).txt
- Errors: 14,000 and counting
- Started over and cleaned up the gene IDs in the excel report so that the names followed the pattern. Ran Expression Dataset Manager again.
- Errors: 1820
- Had to tweak the database with new customizations and re-export.
- 8354 Ordered Locus names in Microsoft Access. All IDS now have two formats LmjF.##.#### and LmjF_##_####.
- Reran Expression Dataset Manager and found 1820 errors.
- The exceptions file was posted to the Leishmania wiki page