Difference between revisions of "Troque Week 12"
From LMU BioDB 2015
(Made Week 12 journal) |
m (→Export Information: Linking vanilla export) |
||
| (14 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{Template:Troque}} | {{Template:Troque}} | ||
| + | {{Template:Oregon Trail Survivors}} | ||
| + | == Things to note == | ||
| + | * Taxonomy ID: 623 | ||
| + | * UP000001006 | ||
| + | * File management system: Wiki | ||
| + | == Export Information == | ||
| + | Version of GenMAPP Builder: | ||
| + | * ''' gmbuilder-3.0.0-build-5 ''' | ||
| + | |||
| + | Computer on which export was run: | ||
| + | * ''' Front of the room, 3rd computer from the right. ''' | ||
| + | |||
| + | Postgres Database name: | ||
| + | * '''Shigella flexneri 20151911''' | ||
| + | |||
| + | UniProt XML filename (give filename and upload and link to compressed file): | ||
| + | * UniProt XML version (The version information can be found at [http://uniprot.org/news the UniProt News Page]): '''UniProt release 2015_11''' | ||
| + | * UniProt XML download link: '''[http://www.uniprot.org/uniprot/?query=proteome:UP000001006 Click here]''' | ||
| + | * Time taken to import: '''4.48 minutes''' | ||
| + | ** Note: | ||
| + | |||
| + | GO OBO-XML filename (give filename and upload and link to compressed file): | ||
| + | * GO OBO-XML version (The version information can be found in the file properties after the file downloaded from the [http://beta.geneontology.org/page/download-ontology GO Download page] has been unzipped): '''Version created on 11/19/2015 (at 2:24 AM)''' | ||
| + | * GO OBO-XML download link: '''[http://archive.geneontology.org/latest-termdb/go_daily-termdb.obo-xml.gz Click here to download].''' | ||
| + | * Time taken to import: '''7.00 minutes''' | ||
| + | * Time taken to process: '''4.99 minutes''' | ||
| + | ** Note: | ||
| + | |||
| + | GOA filename (give filename and upload and link to compressed file): | ||
| + | * GOA version (News on [http://www.ebi.ac.uk/GOA/ this page] records past releases; current information can be found in the Last modified field on the [ftp://ftp.ebi.ac.uk/pub/databases/GO/goa/proteomes/ FTP site]): '''Version released on .''' | ||
| + | * GOA download link: '''[http://ftp.ebi.ac.uk/pub/databases/GO/goa/proteomes/103.S_flexneri_301.goa Click here to download].''' | ||
| + | * Time taken to import: '''0.06 minutes''' | ||
| + | ** Note: | ||
| + | |||
| + | Name of .gdb file (give filename and upload and link to compressed file): '''[[Media:Sf-Std 20151119 OTS.gdb | Sf-Std_20151119_OTS.gdb]]''' | ||
| + | * Time taken to export: ''' 1 Hours, 32 Minutes, 33 Seconds ''' | ||
| + | ** Start time: '''4:06:13 PM PM PDT''' | ||
| + | ** End time: '''5:38:46 PM PDT''' | ||
| + | ** Note: | ||
| + | |||
| + | ==TallyEngine== | ||
| + | |||
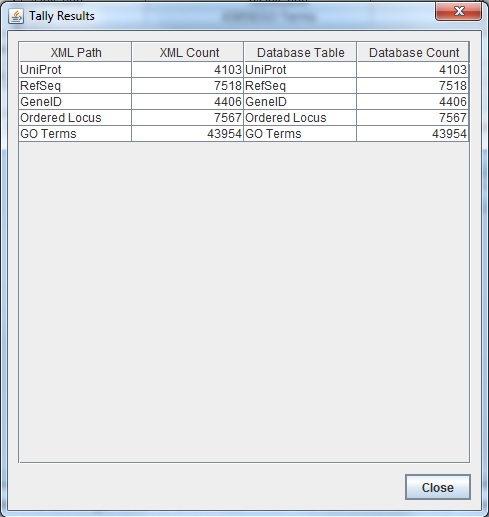
| + | * Run the TallyEngine in GenMAPP Builder and record the number of records for UniProt and GO in the XML data and in the Postgres databases. | ||
| + | ** Choose the menu item Tallies > Run XML and Database Tallies for UniProt and GO... | ||
| + | ** Choose the UniProt and GO OBO XML files that was uploaded from the previous sections of this assignment. | ||
| + | ** Here is the screenshot of the tally result: | ||
| + | <div class="center" style="width: auto; margin-left: auto; margin-right: auto;"> | ||
| + | [[Image:TallyEngine results OTS 112115.jpg]] | ||
| + | </div> | ||
| + | |||
| + | == Using XMLPipeDB match to Validate the XML Results from the TallyEngine== | ||
| + | [[How_Do_I_Count_Thee%3F_Let_Me_Count_The_Ways | Follow the instructions found on this page to run XMLPipeDB match.]] | ||
| + | * In the Thawspace directory, I created a folder called "Shigella_flexneri_BioDB_2015" and created subfolders called "Source" and "Working" to store the source files (i.e., the compressed files) and the working files (i.e., the files I will actually be processing). | ||
| + | * As a result, I had to cd to these directories first before using the command for using Match. | ||
| + | ** In order to change into the ThawSpace0\Shigella_flexneri_BioDB_2015\Working directory, use the following commands on the command prompt window: | ||
| + | T: && cd "Shigella_flexneri_BioDB_2015\Working" | ||
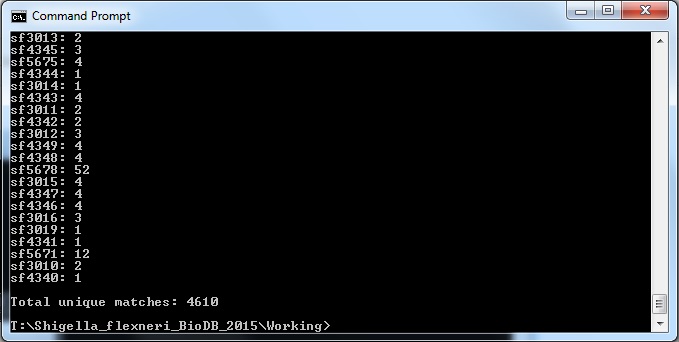
| + | * The command I used once inside the directory I want is: | ||
| + | java -jar xmlpipedb-match-1.1.1/xmlpipedb-match-1.1.1.jar "SF[0-9][0-9][0-9][0-9]" < uniprot-proteome%3AUP000001006.xml | ||
| + | * The results are as follows: | ||
| + | <div class="center"> | ||
| + | [[Image:Match results OTS 112115.jpg]] | ||
| + | </div> | ||
| + | These results did not match up with what the TallyEngine gave (TallyEngine: 7567 vs. Match: 4610) | ||
| + | * As a result, the commands would have to be modified somehow so that the numbers match. | ||
| + | |||
| + | == System IDs == | ||
| + | * UniProt: A0A0H2[A-Z][A-Z][0-9][0-9], A0A0H2[A-Z][A-Z][A-Z][0-9], A0A0H2[A-Z][0-9][0-9][0-9], or [A-Z][0-9][A-Z][0-9][A-Z][0-9] | ||
| + | ** Examples: A0A0H2USI9, A0A0H2USA4, A0A0H2V010, A5A6A8 | ||
| + | * RefSeq: NP_######, WP_#########, YP_#########, YP_###### | ||
| + | ** Examples: NP_858405, WP_000002440, YP_001449236, YP_145811 (only one of this) | ||
| + | * GeneID (EntrezGene from NCBI): | ||
| + | * GO: ####### | ||
| + | * OrderedLocusNames: CP####, S#### (or S####.#), and SF#### (or SF####.#) | ||
{{Template:Troque_Journal}} | {{Template:Troque_Journal}} | ||
Latest revision as of 05:55, 8 December 2015
Contents
Helpful Links
| Team Links | |||||
|---|---|---|---|---|---|
| Files | Team Members | Week 11 Assignment | Week 12 Assignment | Week 14 Assignment | Week 15 Assignment |
| OTS Deliverables | Trixie | Week 11 | Week 12 | Week 14 | Week 15 |
| Jake | Week 11 | Week 12 | Week 14 | Week 15 | |
| Gene Database Testing Report | Erich | Week 11 | Week 12 | Week 14 | Week 15 |
| Kristin | Week 11 | Week 12 | Week 14 | Week 15 | |
| Gene Database Project Links | |||||||
|---|---|---|---|---|---|---|---|
| Overview | Deliverables | Reference Format | Guilds | Project Manager | GenMAPP User | Quality Assurance | Coder |
| Teams | Heavy Metal HaterZ | The Class Whoopers | GÉNialOMICS | Oregon Trail Survivors | |||
Things to note
- Taxonomy ID: 623
- UP000001006
- File management system: Wiki
Export Information
Version of GenMAPP Builder:
- gmbuilder-3.0.0-build-5
Computer on which export was run:
- Front of the room, 3rd computer from the right.
Postgres Database name:
- Shigella flexneri 20151911
UniProt XML filename (give filename and upload and link to compressed file):
- UniProt XML version (The version information can be found at the UniProt News Page): UniProt release 2015_11
- UniProt XML download link: Click here
- Time taken to import: 4.48 minutes
- Note:
GO OBO-XML filename (give filename and upload and link to compressed file):
- GO OBO-XML version (The version information can be found in the file properties after the file downloaded from the GO Download page has been unzipped): Version created on 11/19/2015 (at 2:24 AM)
- GO OBO-XML download link: Click here to download.
- Time taken to import: 7.00 minutes
- Time taken to process: 4.99 minutes
- Note:
GOA filename (give filename and upload and link to compressed file):
- GOA version (News on this page records past releases; current information can be found in the Last modified field on the FTP site): Version released on .
- GOA download link: Click here to download.
- Time taken to import: 0.06 minutes
- Note:
Name of .gdb file (give filename and upload and link to compressed file): Sf-Std_20151119_OTS.gdb
- Time taken to export: 1 Hours, 32 Minutes, 33 Seconds
- Start time: 4:06:13 PM PM PDT
- End time: 5:38:46 PM PDT
- Note:
TallyEngine
- Run the TallyEngine in GenMAPP Builder and record the number of records for UniProt and GO in the XML data and in the Postgres databases.
- Choose the menu item Tallies > Run XML and Database Tallies for UniProt and GO...
- Choose the UniProt and GO OBO XML files that was uploaded from the previous sections of this assignment.
- Here is the screenshot of the tally result:
Using XMLPipeDB match to Validate the XML Results from the TallyEngine
Follow the instructions found on this page to run XMLPipeDB match.
- In the Thawspace directory, I created a folder called "Shigella_flexneri_BioDB_2015" and created subfolders called "Source" and "Working" to store the source files (i.e., the compressed files) and the working files (i.e., the files I will actually be processing).
- As a result, I had to cd to these directories first before using the command for using Match.
- In order to change into the ThawSpace0\Shigella_flexneri_BioDB_2015\Working directory, use the following commands on the command prompt window:
T: && cd "Shigella_flexneri_BioDB_2015\Working"
- The command I used once inside the directory I want is:
java -jar xmlpipedb-match-1.1.1/xmlpipedb-match-1.1.1.jar "SF[0-9][0-9][0-9][0-9]" < uniprot-proteome%3AUP000001006.xml
- The results are as follows:
These results did not match up with what the TallyEngine gave (TallyEngine: 7567 vs. Match: 4610)
- As a result, the commands would have to be modified somehow so that the numbers match.
System IDs
- UniProt: A0A0H2[A-Z][A-Z][0-9][0-9], A0A0H2[A-Z][A-Z][A-Z][0-9], A0A0H2[A-Z][0-9][0-9][0-9], or [A-Z][0-9][A-Z][0-9][A-Z][0-9]
- Examples: A0A0H2USI9, A0A0H2USA4, A0A0H2V010, A5A6A8
- RefSeq: NP_######, WP_#########, YP_#########, YP_######
- Examples: NP_858405, WP_000002440, YP_001449236, YP_145811 (only one of this)
- GeneID (EntrezGene from NCBI):
- GO: #######
- OrderedLocusNames: CP####, S#### (or S####.#), and SF#### (or SF####.#)
Assignment Links
Weekly Assignments
- Week 1
- Week 2
- Week 3
- Week 4
- Week 5
- Week 6
- Week 7
- Week 8
- Week 9
- Week 10
- Week 11
- Week 12
- No Week 13 Assignment
- Week 14
- Week 15
Individual Journal Entries
- Week 1 - This is technically the user page.
- Week 2
- Week 3
- Week 4
- Week 5
- Week 6
- Week 7
- Week 8
- Week 9
- Week 10
- Week 11
- Week 12
- No Week 13 Assignment
- Week 14
- Week 15