Difference between revisions of "Vpachec3 Week 9"
(deleted section) |
(→Links: Added template for journal links) |
||
| Line 189: | Line 189: | ||
==Links== | ==Links== | ||
[https://xmlpipedb.cs.lmu.edu/biodb/fall2015/index.php/User:Vpachec3 Vpachec3 User Page] | [https://xmlpipedb.cs.lmu.edu/biodb/fall2015/index.php/User:Vpachec3 Vpachec3 User Page] | ||
| + | |||
| + | {{Template:Vpachec3 journal links}} | ||
[[Category: Journal Entry]] | [[Category: Journal Entry]] | ||
Latest revision as of 08:00, 4 November 2015
Contents
- 1 October 27, 2015
- 2 October 29,2015
- 3 Export Information
- 4 TallyEngine
- 5 Using XMLPipeDB match to Validate the XML Results from the TallyEngine
- 6 Using SQL Queries to Validate the PostgreSQL Database Results from the TallyEngine
- 7 OriginalRowCounts Comparison
- 8 Visual Inspection
- 9 .gdb Use in GenMAPP
- 10 Links
October 27, 2015
I was able to follow along in class up until I need to go into gmbuilder.bat. It turns out that the computer I was using did not have Java 8 on it. Thus, I wasn't able to continue to work on my computer. However, I was able to see what my partner, Mahrad, was doing on his computer.
The following are the notes taken during class:
2.86 minutes to import the data Uniport XML
6.99 minutes Go OBO XML
4.46 minutes GOA
Start time:3:52 pm
End time: Class time ran out before it was finished, so we are leaving the windows open until next class period.
October 29,2015
When we came back, the computer said the end time was 4:45pm, which was 53 minutes.
The computer used hasn't been updated with Java thus, today I will be working with Mahrad through the protocol.
We went through the protocol for tally engine. All of our numbers matched between the different columns. Mahrad took a screen shot of the table.
We learned about the cmd which talks directly to each particular computer.
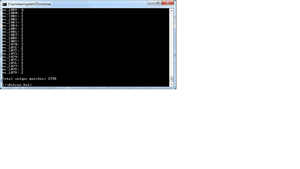
When we ran java -jar xmlpipedb-match-1.1.1.jar "VC_[0-9][0-9][0-9][0-9]" < uniprot-organism%3A243277.xml we got 2738 total unique matches.
This is alarming because the tally engine said the ID's were 3831 but the xmlfile is a much lower. We are going to find the numbers for PGAdminIII and the gdb to see the rest of the numbers.
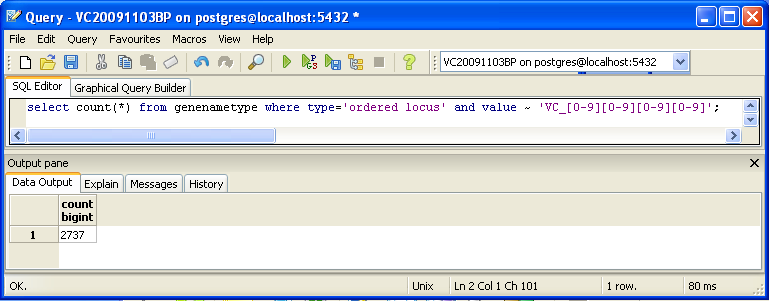
In PGAdminIII we got the number 2737.
In the gdb, we got the number 7664.
Observation from the class:
Some ids start with vca###
Some have underscores while some don't
My partner said that the 7664 is almost double the 3831. Thus, the underscore caused a discrepancy between the numbers.
Needed to ad OR to the SQL and take into account the A in VC_A and the number came out to be 3831.
We need to change the command to java -jar xmlpipedb-match-1.1.1.jar "VC_A?[0-9][0-9][0-9][0-9]" < uniprot-organism%3A243277.xml
Dump into text file:
java -jar xmlpipedb-match-1.1.1.jar "VC_A?[0-9][0-9][0-9][0-9]" < uniprot-organism%3A243277.xml > results.txt
This is where we stopped for the day.
Export Information
Version of GenMAPP Builder: gmbuilder-3.0.0-build-5
Computer on which export was run: SEA120-04
Postgres Database name: Vcholerae_20151027_MS
UniProt XML filename (give filename and upload and link to compressed file):uniport-organism%3A243277.xml
- UniProt XML version (The version information can be found at the UniProt News Page):UniProt release 2015_10
- UniProt XML download link:<http://www.uniprot.org/uniprot/?query=organism:243277>
- Time taken to import: 2.86 minutes
- Note:The computer I was working on did not have Java installed. I worked with my partner Mahrad to complete this assignment.
GO OBO-XML filename (give filename and upload and link to compressed file):go_daily-termdb.obo.xml
- GO OBO-XML version (The version information can be found in the file properties after the file downloaded from the GO Download page has been unzipped):
- GO OBO-XML download link: <http://geneontology.org/page/download-ontology#Legacy_Downloads>
- Time taken to import:6.99 minutes
- Time taken to process:
GOA filename (give filename and upload and link to compressed file): 46.V_cholerae_ATCC_39315.goa
- GOA version (News on this page records past releases; current information can be found in the Last modified field on the FTP site):
- GOA download link:<http://ftp.ebi.ac.uk/pub/databases/GO/goa/proteomes/>
- Time taken to import: 4.46 minutes
Name of .gdb file (give filename and upload and link to compressed file):Vc-Std_20151027_MS.gdb
- Time taken to export:53 mins
- Start time: 3:52 pm
- End time: 4:45 pm
Note: We had to leave the computers after class with a note so that there are no interruptions to the export.
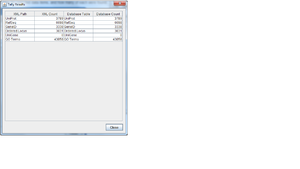
TallyEngine
- Run the TallyEngine in GenMAPP Builder and record the number of records for UniProt and GO in the XML data and in the Postgres databases.
- Choose the menu item Tallies > Run XML and Database Tallies for UniProt and GO...
- Take a screenshot of the results. Upload the image to the wiki and display it on this page.
Using XMLPipeDB match to Validate the XML Results from the TallyEngine
Follow the instructions found on this page to run XMLPipeDB match.
Are your results the same as you got for the TallyEngine? Why or why not?
The results were not the same. They were off by a bit so in class we decided that we are going to check the other programs to see what the numbers were and then come together to brainstorm the possible errors in the process.
This is particularly alarming because the numbers aren't just off by a couple of numbers, there is a large difference between the two.
Using SQL Queries to Validate the PostgreSQL Database Results from the TallyEngine
For more information, see this page.
You can also look for counts at the SQL level, using some variation of a select count(*) query. This requires some knowledge of which table received what data. Here’s an initial tip: the gene/name tags in the XML file land in the genenametype table. A query on this table counting values from this table that were marked as ordered locus in the XML file matching the pattern VC_[0-9][0-9][0-9][0-9] would look like this:
select count(*) from genenametype where type = 'ordered locus' and value ~ 'VC_[0-9][0-9][0-9][0-9]';
In pgAdmin III, you can issue these queries by clicking on the pencil/SQL icon in the toolbar, typing the query into the SQL Editor tab, then clicking on the green triangular Play button to run.
Are your results the same as reported by the TallyEngine? Why or why not?
When Mahrad and I did this in class, we got in PGAdminIII the number 2737. This is again not matching up with the counts. We have one more program to check before we can see compare all numbers and files to try to identify the problem.
OriginalRowCounts Comparison
Within the .gdb file, look at the OriginalRowCounts table to see if the database has the expected tables with the expected number of records. Compare the tables and records with a benchmark .gdb file.
Benchmark .gdb file: Vc-Std_External_20101022.gdb
Copy the OriginalRowCounts table from the benchmark and new gdb and paste them here:In the gdb, we got the number 7664.
Note:
Visual Inspection
Perform visual inspection of individual tables to see if there are any problems.
- Look at the Systems table. Is there a date in the Date field for all gene ID systems present in the database? No,it does not have dates next to all of the databases mentioned.
- Open the UniProt, RefSeq, and OrderedLocusNames tables. Scroll down through the table. Do all of the IDs look like they take the correct form for that type of ID?
We noticed that the ID's were not all written in unison. There were ID's that started with different combinations:
- VC_####
- VC_A####
- VC####
- VCA####
We figured that this would have been the main cause for the discrepancies in the numbers between the different programs. It is important to note that the underscore was put there on purpose therefore must be account for. This observation about the differe ID's would enable us to take the 7664 and divide it by half. However, the numbers are still off by two.
We found that the gene ID, "VC_A0360.1" was causing the problem because it's decimal format in unique to all of the rest of the ID's. Therefore, this is where the extra two were coming from.
After this process, we now finall have the numbers matching.
.gdb Use in GenMAPP
Note:
Putting a gene on the MAPP using the GeneFinder window
- Try a sample ID from each of the gene ID systems. Open the Backpage and see if all of the cross-referenced IDs that are supposed to be there are there.
I first tried putting in the GeneFinder in the Help Window to know where to find it. And also, there was added instruction to the wiki instruction page. I used VC0028 as suggested.
Creating an Expression Dataset in the Expression Dataset Manager
How many of the IDs were imported out of the total IDs in the microarray dataset? How many exceptions were there? Look in the EX.txt file and look at the error codes for the records that were not imported into the Expression Dataset. Do these represent IDs that were present in the UniProt XML, but were somehow not imported? or were they not present in the UniProt XML?
There 121 were errors. All of the errors were labeled “Gene not found in OrderedLocusNames or any related system.” This makes sense since I used the 2010 data last week and there was a 121 error message while my partner had a larger, 727. Also, we searched for that specific error labeled in the data set in last week's assignment as well so this is to be expected. I searched using Acess through the UniProt table for VC2209 which had an error next to it. It was not found in the table, so the ID's that produced errors were not present in the UniProt XML.
Coloring a MAPP with expression data
Note: This was the direction from week 8. I used the same coloring, increased red, decreased green and the same criterion.
Running MAPPFinder
Links
- Individual Journal Week 2
- Individual Journal Week 3
- Individual Journal Week 4
- Individual Journal Week 5
- Individual Journal Week 6
- Individual Journal Week 7
- Individual Journal Week 8
- Individual Journal Week 9
- Individual Journal Week 10
- Individual Journal Week 11
- Individual Journal Week 12
- Individual Journal Week 13
- Individual Journal Week 14
- Individual Journal Week 15