Difference between revisions of "Vpachec3 Week 10"
(→Annotated Bibliography: changed title and deleted a couple lines) |
(→Microarray Paper Search Method: deleted section) |
||
| Line 56: | Line 56: | ||
I noticed that on the right side there was a section that says "Titles with your search terms" and the first paper was the same paper from the Google Scholar search. I would take a that one first. | I noticed that on the right side there was a section that says "Titles with your search terms" and the first paper was the same paper from the Google Scholar search. I would take a that one first. | ||
# At this point, Brandon had already found a paper and as a group we wanted to take a look at it so he posted on the group page. I clicked on the link and it was the golden paper. So we had agreed to use that one as our genome paper. | # At this point, Brandon had already found a paper and as a group we wanted to take a look at it so he posted on the group page. I clicked on the link and it was the golden paper. So we had agreed to use that one as our genome paper. | ||
| − | |||
| − | |||
===Microarray Search Methods=== | ===Microarray Search Methods=== | ||
Revision as of 01:40, 8 November 2015
Contents
Tuesday November 3, 2015
We came together as a team to choose our species, Project Manager and our team name.
Additional Class Notes:
- Genome sequence paper
- Microarray data/paper
- There will be duplicated information on user page and teams page
- Make use of LibGuide through the library; databases section on class's LibGuide page for very useful links
- Use APA style because that is closest to the usual format used in biology
- DOI is a primary id or unique identifier that is specific for each article
- Pubmed Central contains the full text of papers in it, while Pubmed just has abstracts of articles
- Copyright-who owns the work, owns the right to distribute the work
- For microarray, find datasets then find paper- would be easier
Thursday November 5,2015
Demonstration of PubMed
- go straight to advanced search; its the front end of SQL
- use filters to narrow down the search
Demonstration of Google Scholar
- has a wider range
- can see how many times a paper has been cited in other papers
- also has filters to narrow down search
Demonstration of Web of Science
- prospective search: you can see where the paper is being cited; google can do this too
- Web of Science is better for organization and retrieval
Demonstration of ArrayExpression
- filter by experiment type
- high number of arrays are better
So first we needed to decide on a genome sequence paper. Brandon was the first one to find the paper through web of science. He shared it on the group page.
He used the key search as Burkholderia cenocepacia genome and filtered by number of citations. The paper with the highest citations will appear first.
Search Methods
Genome Paper Search Method
The way I decided to approach finding a genome paper was by first finding a data set then use the data set to link us to a paper
- Starting in ArrayExpress, I typed in a a key term the name of the species: Burkholderia cenocepacia
- With that keyword alone, the results were 35 experiments. To narrow down the results, I used the filters where I chose to filter by experiment type which had two drop down bars. The first bar I selected RNA assay and the second bar I selected Array assay. This led to having 24 experiments.
- Within this search, my teammates made a note that looking through the titles, there were experiments using different strains. We decided to use the J2315 strain.
- Adding the J2315 into the keyword search, narrowed down the experiment results to 16.
- To select through the 16, we are looking at the assays column. We want a number greater than or equal to 9 to ensure better quality data.
- I took a took a title from the results and used in the other search engines. But then I got confused and didn't really understand what the next step would be. So, I just started to use the other search engines independently.
- Looking at Web of Science, I typed in the key term Burkholderia cenocepacia genome.
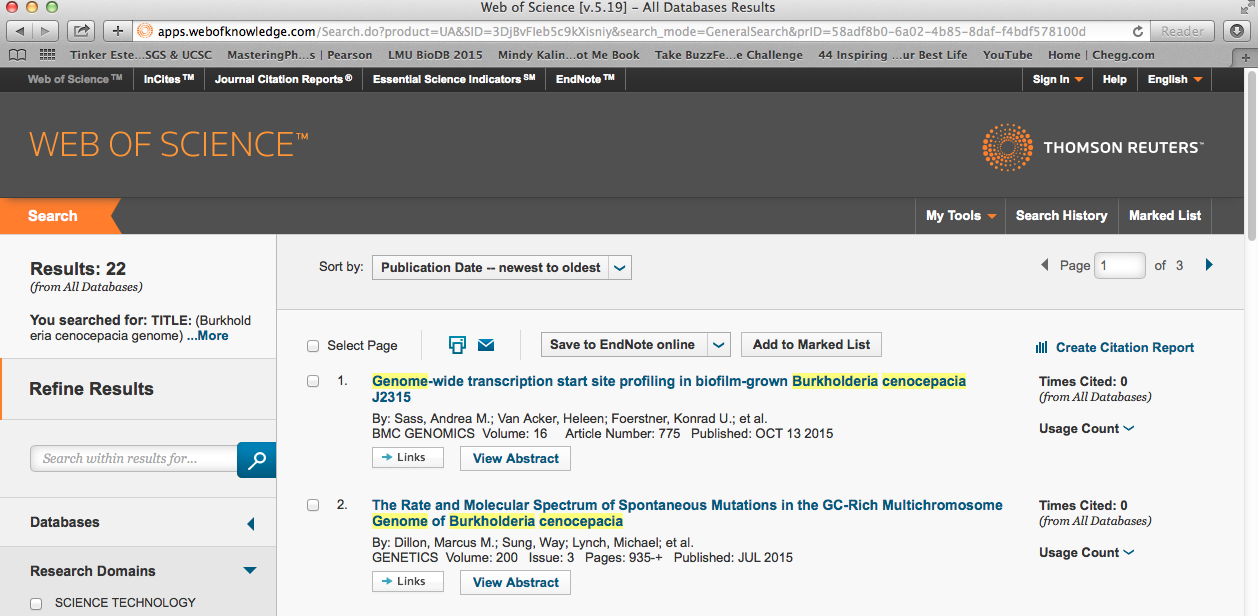
There were 22 results. Before adding any filter, I want to read through the first five papers to get an idea of how to filter. The first paper was recent, Oct 13 2015 and hasn't been cited. However, it was looking at the particular strain we want: J2315.
- I opened another tab and used google scholar. Again, I typed in the same key term just to get an idea of how to filter based on the first few results.
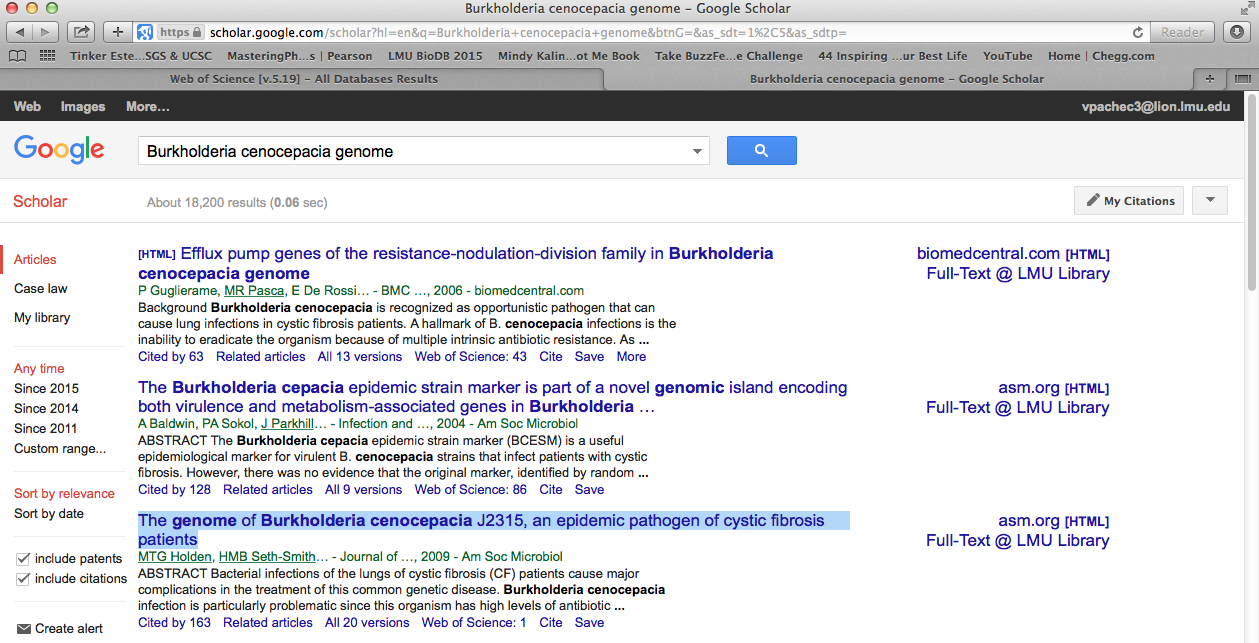
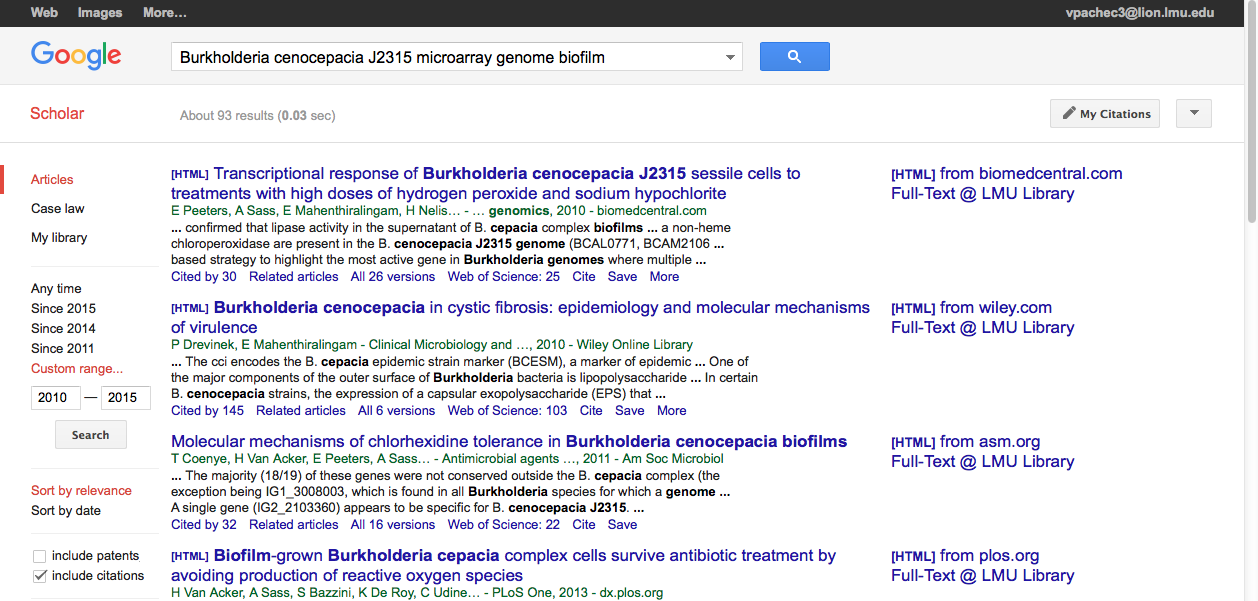
There were 18,200 results. The first two didn't seem particularly helpful and they were past or almost past the 10 year mark. However, the third article looked golden. The title was looked like the paper would have the genome data needed, it was within the 10 year mark, it was cited 163 times and the full text could be found at the LMU library. This one I was definitely going to come back to.
I noticed that on the right side there was a section that says "Titles with your search terms" and the first paper was the same paper from the Google Scholar search. I would take a that one first.
- At this point, Brandon had already found a paper and as a group we wanted to take a look at it so he posted on the group page. I clicked on the link and it was the golden paper. So we had agreed to use that one as our genome paper.
Microarray Search Methods
- Use a keyword search for each of these databases/tools and answer the following:
- PubMed
- What original keyword(s) did you use? How many results did you get? I used the keywords Burkholderia cenocepacia J2315 microarray and got 4 results
- Which terms in which combinations were most useful to narrow down the search? How many results did you get after narrowing the search? I deleted the key term microarray to see how many results I would get and it got 67 hits. So the term microarray narrowed down the search very much. I also added a filter for the free full text because it would be what I need to read the article. And after that I got 3 hits. However, the one with was taken away actually seems like a pretty good candidate for an article so I am going to save that and see if I can find the full text elsewhere.
- PubMed
- Use a keyword search for each of these databases/tools and answer the following:
- Google Scholar
- What original keyword(s) did you use? How many results did you get? I used the keywords Burkholderia cenocepacia J2315 microarray. Since Google has a wider range, there 210 results.
- Which terms in which combinations were most useful to narrow down the search? How many results did you get after narrowing the search? To narrow down the search, I filtered by time range. I had a 5 year limit from 2005 to 2015 and this brought the hits down to 191. I then added the key terms biofilm and genome. This narrowed down the search to 93. I also unchecked the patents but it did not affect the number.
- Google Scholar
- Web of Science
- What original keyword(s) did you use? How many results did you get? I started off the the full search:Burkholderia cenocepacia J2315 microarray genome biofilm and it only got 2 hits.
- Which terms in which combinations were most useful to narrow down the search? How many results did you get after narrowing the search? I reentered the search without the terms genome and biofilm. I also added the time span to ten years from 2005 to 2015. This got me 32 results. After I added the key term biofilm and it went down to 21 hits.
- Web of Science
- Use the advanced search functions for each of these three databases/tools and answer the following:
- PubMed
- Which advanced search functions were most useful to narrow down the search? How many results did you get?Since the search that resulted in 4 hits, I wasn't able to narrow down the search even further. However, in the advanced section, it was pretty neat to see that the web site save my search histories so you can see the thought process and the actions to get to the final search.
- PubMed
- Use the advanced search functions for each of these three databases/tools and answer the following:
- Google Scholar
- Which advanced search functions were most useful to narrow down the search? How many results did you get? In the advanced search option, I was able to arrow down the search by saying I just wanted my key terms in my titles. I also deleted the word microarray. It got it down to 26.
- Google Scholar
- Web of Science
- Which advanced search functions were most useful to narrow down the search? How many results did you get? Using the advanced search option, I was able to type in my keywords a bit differently. I used the formula TS=(Burkholderia cenocepacia J2315 AND microarray) and also refined by using dataset documents only. This got 16 hits. The datasets linked back to the abstracts so I can find the relating articles through there.
- Web of Science
- Each of the references in your bibliography needs to have the following information (an example is given in another section below):
- The complete bibliographic reference in the APA style (see the Writing LibGuide) You will be using one of three formats, “journal article from database (with DOI), journal article from database (no DOI) or journal article in print (no DOI).)
- The link to the abstract from PubMed.
- The link to the full text of the article in PubMedCentral.
- The link to the full text of the article (HTML format) from the publisher web site.
- The link to the full PDF version of the article from the publisher web site.
- Who owns the rights to the article?
- Does the journal own the copyright?
- Do the authors own the copyright?
- Do the authors own the rights under a Creative Commons license?
- Is the article available “Open Access”?
- What organization is the publisher of the article? What type of organization is it? (commercial, for-profit publisher, scientific society, respected open access organization like Public Library of Science or BioMedCentral, or predatory open access organization, see the list of) (Open Access Scholarly Publishers Association Members) here.
- Is this article available in print or online only?
- Has LMU paid a subscription or other fee for your access to this article?
- Each of the references in your bibliography needs to have the following information (an example is given in another section below):
- Use the genome sequencing article you found to perform a prospective search in the ISI Web of Science/Knowledge database.
- Give an overview of the results of the search.
- How many articles does this article cite?
- How many articles cite this article?
- Based on the titles and abstracts of the papers, what type of research directions have been taken now that the genome for that organism has been sequenced?
- Give an overview of the results of the search.
- Each person needs to find 1-2 potential journal articles that refer to public/published microarray data for your species than are different than what your teammates have found. Thus, each team should find 4-8 articles. If you cannot find a minimum of four articles, please let the instructors know right away.
- The experiments must be measuring gene expression aka transcriptional profiling or transcription profiling by array. Microarrays can also be used for other types of experiments, but these won't be suitable for analysis.
- We recommend that you begin by searching for the data, and then by finding the journal article related to the data. State which database you used to find the data and article.
- State what you used as search terms and what type of search terms they were.
- Give an overview of the results of the search.
- How many results did you get?
- Give an assessment of how relevant the results were.
- For each article, please provide all of the same information that you provided for the genome article above.
- In addition, you must also link to the web site where the microarray data resides.
- For each of the microarray articles/datasets, answer the following:
- What experiment was performed? What was the "treatment" and what was the "control" in the experiment?
- Were replicate experiments of the "treatment" and "control" conditions conducted? Were these biological or technical replicates? How many of each?
- Remember, microarray data is not centrally located on the web. Some major sources are:
- EBI ArrayExpress (recommended)
- Click on the link to "Browse ArrayExpress"
- Use the drop down "Filter Search Results" to filter datasets by your organism, by "RNA assay" and "Array assay" to narrow your search.
- NCBI GEO
- Stanford Microarray Database
- PUMAdb (Princeton Microarray Database)
- In addition, microarray data can sometimes be found as supplementary information with a journal article or on an investigator's own web site.
- EBI ArrayExpress (recommended)
- For each of the microarray articles/datasets, answer the following:
- On your team wiki page, compile the list of citations, links, and answers to questions, ranking the papers one through eight in order of preference for using the dataset for your project. The instructors will review your results to make sure that the data are suitable for the project before you move forward with the analysis.