Difference between revisions of "Kevin Wyllie Week 12"
From LMU BioDB 2015
(Syntax fix.) |
(Added a to do list.) |
||
| Line 21: | Line 21: | ||
#*# Then, rows 4-14 were deleted. ''Note: these row numbers are only accurate when these deletions are performed in order.'' | #*# Then, rows 4-14 were deleted. ''Note: these row numbers are only accurate when these deletions are performed in order.'' | ||
#* Finally, a new row was inserted under the header row. These row was titled "ExpName". The purpose of this row is to indicate what kind of cells were used for the corresponding experiment. Cells B2-F2 contain "Biofilm" as these columns correspond to experiments using biofilm cells that were ''not'' treated with tobramycin (125_1, 125_2, 125_3, 125_4, 126_1). Cells G2-I2 contained "Tobramycin" as these columns correspond to experiments using biofilm cells that were treated with tobramycin (126_2, 126_3, 126_4). | #* Finally, a new row was inserted under the header row. These row was titled "ExpName". The purpose of this row is to indicate what kind of cells were used for the corresponding experiment. Cells B2-F2 contain "Biofilm" as these columns correspond to experiments using biofilm cells that were ''not'' treated with tobramycin (125_1, 125_2, 125_3, 125_4, 126_1). Cells G2-I2 contained "Tobramycin" as these columns correspond to experiments using biofilm cells that were treated with tobramycin (126_2, 126_3, 126_4). | ||
| + | |||
| + | === To Do === | ||
| + | |||
| + | * Begin statistical analysis. | ||
| + | * Upload spreadsheet. | ||
| + | * Add good-practice content/links. | ||
Revision as of 20:50, 23 November 2015
Electronic Lab Notebook
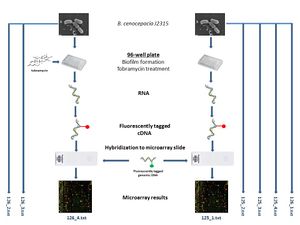
Sample-Data Relationship:
Compiling Data
- From each raw individual data file, columns M ("GeneName") and R ("LogRatio") were copied and pasted into a new Excel file.
- All of the GeneName columns were pasted adjacently (columns A-H).
- Similarly, all LogRatio columns were pasted adjacently (columns I-P).
- To rule out the possibility of confusing any two files with each other, a header was added at the top of each column, with each corresponding file name for example (125_2, for example).
- The GeneName columns were scanned for any discrepancies n terms of amount of rows or ordering of gene ID's. The LogRatio columns were scanned for discrepancies in amount of rows.
- This sheet was named "allgenename_logratio".
- A new sheet was created, named "genename_logratio"
- Only one GeneName column was used for this sheet (column A), as all of the columns had been confirmed to be identical between files.
- The LogRatio data for each file was pasted into columns B through I, maintaining the previously mentioned file name headers. The data was pasted in the order of the file name numbers, from smallest to largest.
- A new sheet was created, named "genename_logratio_clean".
- All content from the "genename_logratio" sheet was pasted into the "genename_logratio_clean" sheet.
- All undesired information was removed.
- First, rows 2-10 were deleted.
- Then, rows 4-14 were deleted. Note: these row numbers are only accurate when these deletions are performed in order.
- Finally, a new row was inserted under the header row. These row was titled "ExpName". The purpose of this row is to indicate what kind of cells were used for the corresponding experiment. Cells B2-F2 contain "Biofilm" as these columns correspond to experiments using biofilm cells that were not treated with tobramycin (125_1, 125_2, 125_3, 125_4, 126_1). Cells G2-I2 contained "Tobramycin" as these columns correspond to experiments using biofilm cells that were treated with tobramycin (126_2, 126_3, 126_4).
To Do
- Begin statistical analysis.
- Upload spreadsheet.
- Add good-practice content/links.