Difference between revisions of "Gene Database Testing Report- cw20151201"
(made 1201 edits) |
(added section on confirming the GenMAPP Updates) |
||
| Line 117: | Line 117: | ||
** Elapsed time: 54 minutes | ** Elapsed time: 54 minutes | ||
Note: No interruptions occurred during the export process. | Note: No interruptions occurred during the export process. | ||
| + | |||
| + | ===Confirming that Changes to GenMAPP Builder Worked As Expected=== | ||
| + | #Systems Table Update | ||
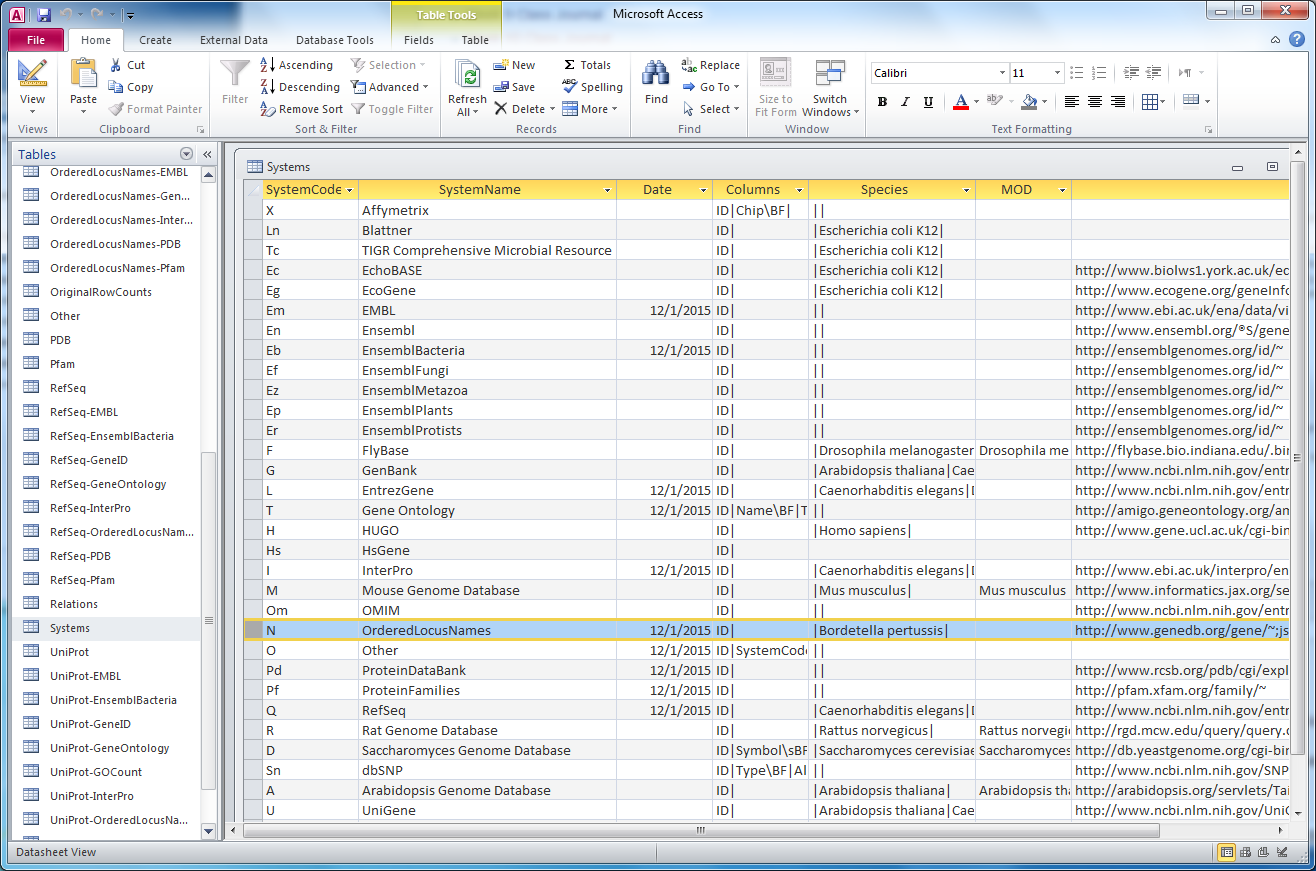
| + | #*In creating the ''Bordetella pertussis'' custom class, changes were implemented to edit the ''OrderedLocusNames'' column in the exported .gdb file. Specifically, the species should be listed as ''Bordetella pertussis'' and a custom GeneDB link should be included in this row. | ||
| + | #*To verify that these changes were working as expected, I opened the file bpertussis-std_cw20151201.gdb in Microsoft Access. Upon opening ''Systems'' table here, all changes were successfully implemented: | ||
| + | #**[[File: Systemstable_cw20151201.png]] | ||
| + | #GenMAPP Gene Backpage Update | ||
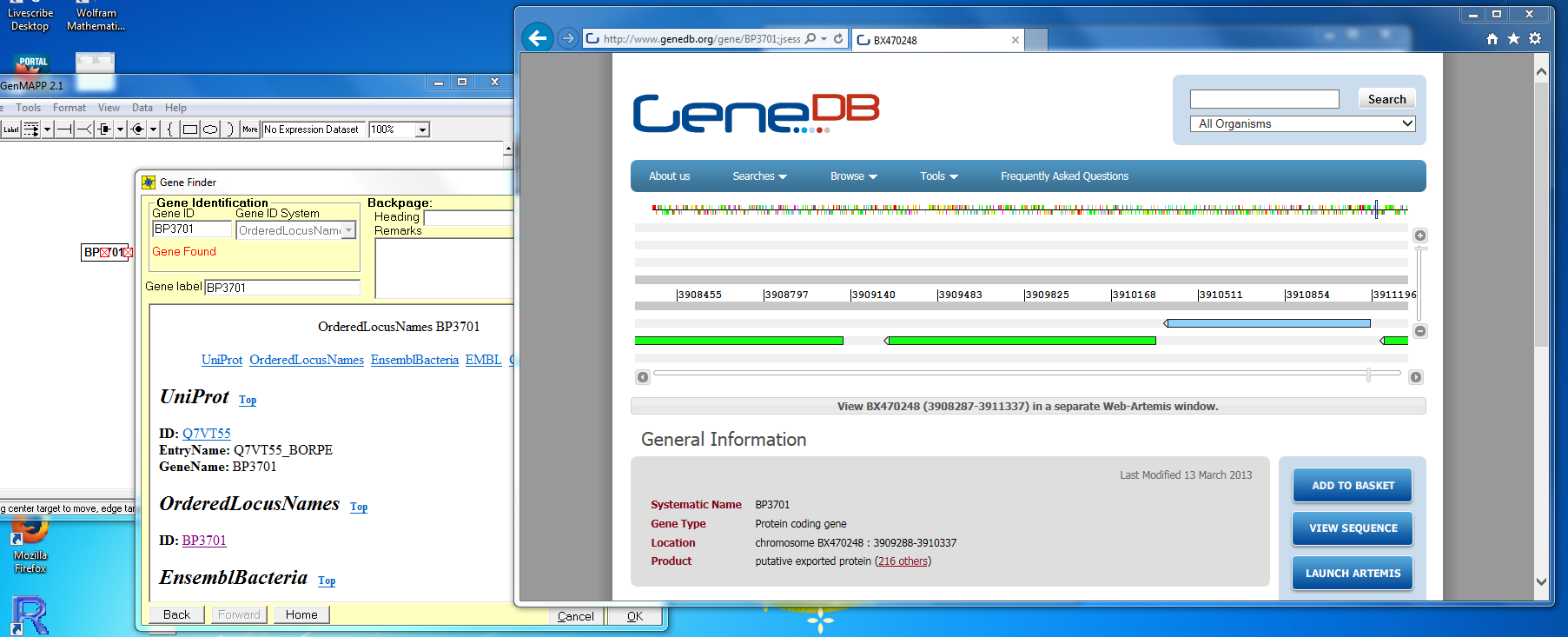
| + | #*The ''OrderedLocusNames'' GeneDB link added (as described above) also was designed to have funcitonality in GenMAPP. Sepcifically, gene backpages opened while using this gene database in GenMAPP should redirect users to the GeneDB information page for the specific ''Bordetella pertussis'' gene. | ||
| + | #*I specified bpertussis-std_cw20151201.gdb as the gene database in GenMAPP and put the gene BP3701 on the map. When I clicked on the link to the ID "BP3701" under "OrderedLocusNames" in Gene Finder, I was successfully brought to the corresponding GeneDB information page: | ||
| + | #**[[File:Custombackpage cw20151201.PNG]] | ||
[[Category: Class Whoopers]] | [[Category: Class Whoopers]] | ||
Revision as of 00:32, 4 December 2015
Contents
- 1 Files Asked for in the Gene Database Testing Report
- 2 Pre-requisites
- 3 Gene Database Creation
- 4 Gene Database Testing Report
Files Asked for in the Gene Database Testing Report
For convenience, all of the files explicitly asked for in the "Gene Database Testing Report" section were compressed together in this file: File:Gdtr cw20151201.zip
Pre-requisites
The following set of software was used in the creation and testing of the Bordetella pertussis gene database:
- 7-ziptool that for unpacking .gz and .zip files
- PostgreSQL on Windows (version 9.4.x)
- GenMAPP Builder
- Java JDK 1.8 64-bit
- GenMAPP 2
- XMLPipeDB match utility for counting IDs in XML files
- Microsoft Access for reading .mdb files
Gene Database Creation
Downloading Data Source Files and GenMAPP Builder
- I download the UniProt XML, GOA, and GO OBO-XML files for Bordetella Pertussis along with the GenMAPP Builder program.
- All files were saved to the folder Bklein7_CW\bpertussis_cw20151201 on my computer's ThawSpace.
- Files that required extraction were unzipped using 7-zip.
- Data files that remained in a folder after unzipping were removed from their folders to facilitate organization and command line processing.
UniProt XML
- I went to the UniProt Complete Proteomes page.
- From there, I navigated to the complete proteome download page for Bordetella pertussis (strain Tohama I / ATCC BAA-589 / NCTC 13251).
- I clicked on the "Download" button at the top of the page above and selected the following options:
- "Download all"
- "XML" from the "Format" drop-down menu
- "Compressed" format
- I extracted the file using 7-zip.
GOA
- UniProt-GOA files can be downloaded from the UniProt-GOA ftp site.
- Within the above site, I navigated to the for Bordetella pertussis strain Tohama I.
- This text file was automatically opened by my browser. Therefore, I had to manually download the file.
GO OBO-XML
- I downloaded the GO OBO-XML formatted file from the Gene Ontology legacy download page.
- I extracted the file using 7-zip.
Downloaded GenMAPP Builder
- I downloaded the first custom version of gmbuilder including the Bordetella pertussis custom class: File:Dist cw20151201.zip.
- I extracted the GenMAPP Builder folder using 7-zip.
Creating the New Database in PostgreSQL
- I launched pgAdmin III and connected to the PostgreSQL 9.4 server (localhost:5432).
- On this server, I created a new database: bpertussis_cw20151201_gmb3build5.
- I opened the SQL Editor tab to use an XMLPipeDB query to create the tables in the database.
- I clicked on the Open File icon and selected the file gmbuilder.sql. This imported a series of SQL commands into the editor tab.
- I clicked on the Execute Query icon to run this command.
- In viewing the schema for this database, I confirmed that there were 167 tables after running the above command.
Configuring GenMAPP Builder to Connect to the PostgreSQL Database
- To begin, I launched gmbuilder.bat.
- I selected the "Configure Database" option and entered the following information into the fields below:
- Host or address: localhost
- Port number: 5432
- Database name: bpertussis_cw20151201_gmb3build5custom
- Username: postgres
- Password: Welcome1
Importing Data into the PostgreSQL Database
- The downloaded data files for Bordetella pertussis were specified and imported into the database by clicking on the following buttons:
- Selected File > Import UniProt XML...
- Selected File > Import GO OBO-XML...
- Clicked OK to the message asking to process the GO data.
- Selected File > Import GOA...
Exporting a GenMAPP Gene Database (.gdb)
- I selected File > Export to GenMAPP Gene Database... to begin the export process.
- I typed my name in the owner field (Brandon Klein).
- I selected "Bordetella pertussis (strain Tohama I/ATCC BAA-589/NCTC 13251), Taxon ID 257313" as the gene database species and then clicked Next.
- The database was saved as bpertussis-std_cw20151201.
- I checked the boxes for exporting all Molecular Function, Cellular Component, and Biological Process Gene Ontology Terms.
- Finally, I clicked the "Next" button to begin the export process.
Gene Database Testing Report
Export Information
Version of GenMAPP Builder: Version 3.0.0 Build 5
Computer on which export was run: Seaver 120- Last computer on the right in the row farthest from the front of the room
Postgres Database name: bpertussis_cw20151201_gmb3build5custom
UniProt XML filename: File:Uniprot-proteome-UP000002676 cw20151201.zip
- UniProt XML version (The version information was found at the UniProt News Page): 2015_11
- UniProt XML download link: Bordetella pertussis (strain Tohama I / ATCC BAA-589 / NCTC 13251)
- Time taken to import: 2.59 minutes
- Note: The import time was nearly equivalent to that when creating the previous "Bordetella pertussis" gene database: bpertussis-std_cw20151119.gdb (2.60 minute). No interruptions occurred during this process.
GO OBO-XML filename: File:Go daily-termdb cw20151201.zip
- GO OBO-XML version (The version information was found in the file properties): Last Modified- December 01, 2015, 2:21:31 AM
- GO OBO-XML download link: Gene Ontology legacy download page
- Time taken to import: 7.08 minutes
- Time taken to process: 4.42 minutes
- Note: The import and processing times were similar to those for the previous "Bordetella pertussis" gene database: bpertussis-std_cw20151119.gdb (6.99 minutes and 4.48 minutes respectively). No interruptions occurred during these processes.
GOA filename: File:145.B pertussis ATCC BAA-589 cw20151201.zip
- GOA version (found in the Last modified field on the FTP site): Last Modified- 11/10/15 1:39:00 PM
- GOA download link: for Bordetella pertussis strain Tohama I
- Time taken to import: 0.04 minutes
- Note: The import time was equal to that of the previous "Bordetella pertussis" gene database: bpertussis-std_cw20151119.gdb. No interruptions occurred during this process.
Name of .gdb file: File:Bpertussis-std cw20151201.zip
- Time taken to export:
- Start time: 4:06 PM
- End time: 5:00 PM
- Elapsed time: 54 minutes
Note: No interruptions occurred during the export process.
Confirming that Changes to GenMAPP Builder Worked As Expected
- Systems Table Update
- In creating the Bordetella pertussis custom class, changes were implemented to edit the OrderedLocusNames column in the exported .gdb file. Specifically, the species should be listed as Bordetella pertussis and a custom GeneDB link should be included in this row.
- To verify that these changes were working as expected, I opened the file bpertussis-std_cw20151201.gdb in Microsoft Access. Upon opening Systems table here, all changes were successfully implemented:
- GenMAPP Gene Backpage Update
- The OrderedLocusNames GeneDB link added (as described above) also was designed to have funcitonality in GenMAPP. Sepcifically, gene backpages opened while using this gene database in GenMAPP should redirect users to the GeneDB information page for the specific Bordetella pertussis gene.
- I specified bpertussis-std_cw20151201.gdb as the gene database in GenMAPP and put the gene BP3701 on the map. When I clicked on the link to the ID "BP3701" under "OrderedLocusNames" in Gene Finder, I was successfully brought to the corresponding GeneDB information page: