Difference between revisions of "Kzebrows Week 9"
(Importing data into PostgreSQL and export) |
(Export of GDB file notes) |
||
| Line 84: | Line 84: | ||
Name of .gdb file (give filename and upload and link to compressed file): | Name of .gdb file (give filename and upload and link to compressed file): | ||
| − | * Time taken to export: | + | * Time taken to export: N/A |
| − | ** Start time: | + | ** Start time: October 27, 2015 at 3:55 pm |
| − | ** End time: | + | ** End time: N/A |
| − | Note: | + | Note: I left a note on top of my computer but someone came in and exited out of my windows between Tuesday's class and Thursday's class, so I was unable to record the end time or time taken to export. The average time of the people around me was around 5:11 pm on Tuesday, so I can infer that my file would have finished exporting around the same time. |
==TallyEngine== | ==TallyEngine== | ||
Revision as of 21:43, 29 October 2015
Contents
- 1 Electronic Lab Notebook
- 2 Export Information
- 3 Create New Database in PostgreSQL
- 4 Configuring GenMAPP Builder to Connect to PostgreSQL Database
- 5 Exporting a GenMAPP Gene Database (.gdb)
- 6 Gene Database Testing Report
- 7 TallyEngine
- 8 Using XMLPipeDB match to Validate the XML Results from the TallyEngine
- 9 Using SQL Queries to Validate the PostgreSQL Database Results from the TallyEngine
- 10 OriginalRowCounts Comparison
- 11 Visual Inspection
- 12 .gdb Use in GenMAPP
- 13 Compare Gene Database to Outside Resource
- 14 Assignments
- 15 Additional Links
Electronic Lab Notebook
Export of Vibrio cholerae GenMAPP Gene Database was used following the instructions on the Running GenMAPP Builder page. The Gene Database Testing Report template we used was found here.
Export Information
First we downloaded UniProt XML, GOA, and GO OBO-XML files. UniProt (protein database) is linked to GO OBO-XML (Gene Ontology) through GOA (Gene Ontology Associations) through XMLPipeDB, subset GenMAPP Builder. GenMAPP Builder takes the data and puts it into PostgreSQL database, where it is converted into a GenMAPP-compatible gene database (GDB). From there, we can then download and analyze microarray data via GenMAPP.
UniProt XML
- Clicked on the UniProt link for Vibrio cholerae serotype O1 and clicked download all with XML format as a compressed file.
- Saved
uniprot-organism%3A243277.xml.gzin Computer T drive under new folderKzebrows - Clicked on the link to download the proteomes directory
- Downloaded file and saw that V_cholerae was downloaded 13 Oct 20016 at 07:31
GOA
- Followed link to Legacy Downloads
- Download obo-xml.gz to T drive Kzebrows
- Downloaded gmbuilder-3.0 and extracted in T drive using 7 zip
Create New Database in PostgreSQL
- Launched PgAdmin III.
- Double-clicked on PostgreSQL 9.4
- Selected "Database" and "New Database" and named it "Vcholerae_20151027_gmb3build5". Copied name to clipboard and pressed OK.
- Ran prepackaged query: open file > Thaw space > gmbuilder-3.0.0-build-5 > sql > gmbuildersql
- Query returned successfully with no results in 5697 ms
- Closed query window
Configuring GenMAPP Builder to Connect to PostgreSQL Database
- Launched gmbuilder.bat
- Select File > Configure Database
- Entered the following info and clicked OK
- Host: localhost
- Port number: 5432
- Database name: Vcholerae_20151027_gmb3build5
- Username: postgres
- password
- File > Import UniProt XML and found UniProt XML file
- File > Import GO OBO-XML file
Exporting a GenMAPP Gene Database (.gdb)
- Selected File > Export to database
- Typed my name into the Owner Field
- Clicked on species V. cholerae
- Created the database by saving under T drive
- Left the boxes checked for exporting all Molecule Function, Cellular Component, and Gene Ontology terms
- Clicked next to begin export process
- Start time: October 27, 2015 at 3:55 pm
Gene Database Testing Report
Version of GenMAPP Builder: gmbuilder-3.0.0-build-5.zip
Computer on which export was run:
Postgres Database name: Vcholerae_20151027_gmb3build5
UniProt XML filename (give filename and upload and link to compressed file):
- UniProt XML version (The version information can be found at the UniProt News Page): 13 Oct 20016 at 07:31
- UniProt XML download link: http://www.uniprot.org/uniprot/?query=organism:243277
- Time taken to import: 3.10 minutes
- Note: Data downloaded slowly.
GO OBO-XML filename (give filename and upload and link to compressed file):
- GO OBO-XML version (The version information can be found in the file properties after the file downloaded from the GO Download page has been unzipped): October 27, 2015 at 3:11 pm
- GO OBO-XML download link: http://archive.geneontology.org/latest-termdb/go_daily-termdb.obo-xml.gz
- Time taken to import: 7.14 minutes
- Time taken to process: 4.64 minutes
- Note: Importing the data took a long time.
GOA filename (give filename and upload and link to compressed file):
- GOA version (News on this page records past releases; current information can be found in the Last modified field on the FTP site):
- GOA download link: http://ftp.ebi.ac.uk/pub/databases/GO/goa/proteomes/46.V_cholerae_ATCC_39315.goa
- Time taken to import: 0.06 minutes
- Note: Import was almost immediate.
Name of .gdb file (give filename and upload and link to compressed file):
- Time taken to export: N/A
- Start time: October 27, 2015 at 3:55 pm
- End time: N/A
Note: I left a note on top of my computer but someone came in and exited out of my windows between Tuesday's class and Thursday's class, so I was unable to record the end time or time taken to export. The average time of the people around me was around 5:11 pm on Tuesday, so I can infer that my file would have finished exporting around the same time.
TallyEngine
- Run the TallyEngine in GenMAPP Builder and record the number of records for UniProt and GO in the XML data and in the Postgres databases.
- Choose the menu item Tallies > Run XML and Database Tallies for UniProt and GO...
- Take a screenshot of the results. Upload the image to the wiki and display it on this page.
- For more information, see this page.
Using XMLPipeDB match to Validate the XML Results from the TallyEngine
Follow the instructions found on this page to run XMLPipeDB match.
Are your results the same as you got for the TallyEngine? Why or why not?
Using SQL Queries to Validate the PostgreSQL Database Results from the TallyEngine
For more information, see this page.
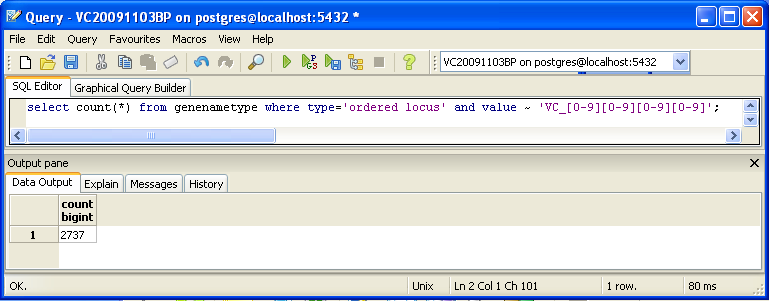
You can also look for counts at the SQL level, using some variation of a select count(*) query. This requires some knowledge of which table received what data. Here’s an initial tip: the gene/name tags in the XML file land in the genenametype table. A query on this table counting values from this table that were marked as ordered locus in the XML file matching the pattern VC_[0-9][0-9][0-9][0-9] would look like this:
select count(*) from genenametype where type = 'ordered locus' and value ~ 'VC_[0-9][0-9][0-9][0-9]';
In pgAdmin III, you can issue these queries by clicking on the pencil/SQL icon in the toolbar, typing the query into the SQL Editor tab, then clicking on the green triangular Play button to run.
Are your results the same as reported by the TallyEngine? Why or why not?
OriginalRowCounts Comparison
Within the .gdb file, look at the OriginalRowCounts table to see if the database has the expected tables with the expected number of records. Compare the tables and records with a benchmark .gdb file.
Benchmark .gdb file:
Copy the OriginalRowCounts table from the benchmark and new gdb and paste them here:
Note:
Visual Inspection
Perform visual inspection of individual tables to see if there are any problems.
- Look at the Systems table. Is there a date in the Date field for all gene ID systems present in the database?
- Open the UniProt, RefSeq, and OrderedLocusNames tables. Scroll down through the table. Do all of the IDs look like they take the correct form for that type of ID?
Note:
.gdb Use in GenMAPP
Note:
Putting a gene on the MAPP using the GeneFinder window
- Try a sample ID from each of the gene ID systems. Open the Backpage and see if all of the cross-referenced IDs that are supposed to be there are there.
Note:
Creating an Expression Dataset in the Expression Dataset Manager
- How many of the IDs were imported out of the total IDs in the microarray dataset? How many exceptions were there? Look in the EX.txt file and look at the error codes for the records that were not imported into the Expression Dataset. Do these represent IDs that were present in the UniProt XML, but were somehow not imported? or were they not present in the UniProt XML?
Note:
Coloring a MAPP with expression data
Note:
Running MAPPFinder
Note:
Compare Gene Database to Outside Resource
The OrderedLocusNames IDs in the exported Gene Database are derived from the UniProt XML. It is a good idea to check your list of OrderedLocusNames IDs to see how complete it is using the original source of the data (the sequencing organization, the MOD, etc.) Because UniProt is a protein database, it does not reference any non-protein genome features such as genes that code for functional RNAs, centromeres, telomeres, etc.
Note:
Assignments
Individual Journal Assignment Pages
- Week 1
- Week 2
- Week 3
- Week 4
- Week 5
- Week 6
- Week 7
- Week 8
- Week 9
- Week 10
- Week 11
- Week 12
- Week 14
- Week 15
Individual Journal Assignments
- Kzebrows Week 1
- Kzebrows Week 2
- Kzebrows Week 3
- Kzebrows Week 4
- Kzebrows Week 5
- Kzebrows Week 6
- Kzebrows Week 7
- Kzebrows Week 8
- Kzebrows Week 9
- Kzebrows Week 10
- Kzebrows Week 11
- Kzebrows Week 12
- Kzebrows Week 14
- Kzebrows Week 15
- Final Individual Reflection
- Class Journal Week 1
- Class Journal Week 2
- Class Journal Week 3
- Class Journal Week 4
- Class Journal Week 5
- Class Journal Week 6
- Class Journal Week 7
- Class Journal Week 8
- Class Journal Week 9
- Oregon Trail Survivors Week 10
- Oregon Trail Survivors Week 11
- Oregon Trail Survivors Week 12
- Oregon Trail Survivors Week 14
Additional Links
- User Page: Kristin Zebrowski
- Class Page: BIOL/CMSI 367-01
- Team Page: Oregon Trail Survivors