Jwoodlee Week 15
Contents
Electronic Lab Notebook
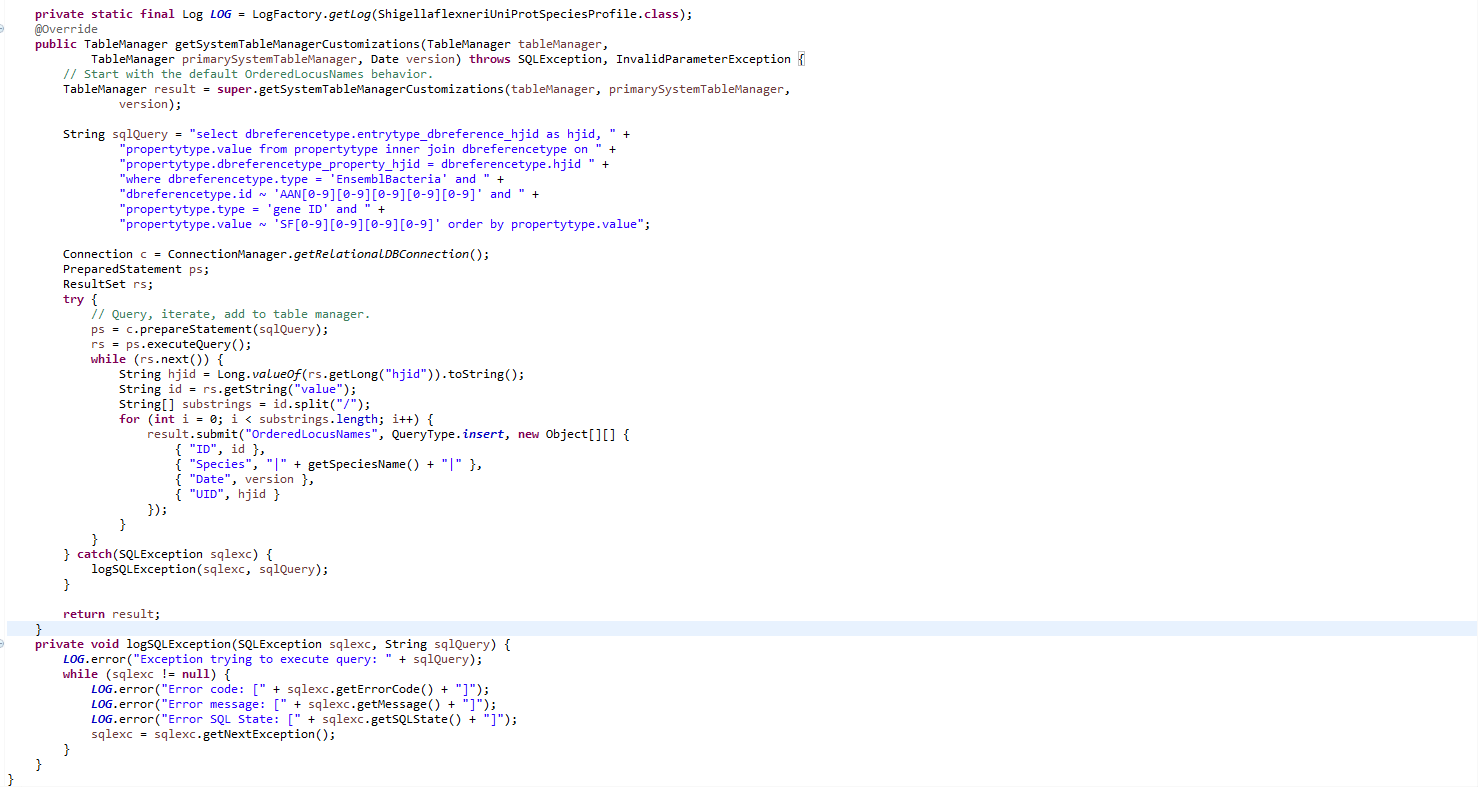
Our GenMAPP users were reporting 416 missing genes from the .gdb which was a problem. Trixie found that 92 of these genes were in the XML file in such a way that led to the exporter missing them, the rest of the missing genes simply weren't in the XML file. Specifically, the genes were somewhere else in the file and weren't added to the OrderedLocusNames table by default. To capture these 92 elusive genes we consulted Dondi. Dondi edited the ShigellaflexneriUniProtSpeciesProfile class, which I had previously constructed, by adding a SQL query that captured the 92 missing genes:
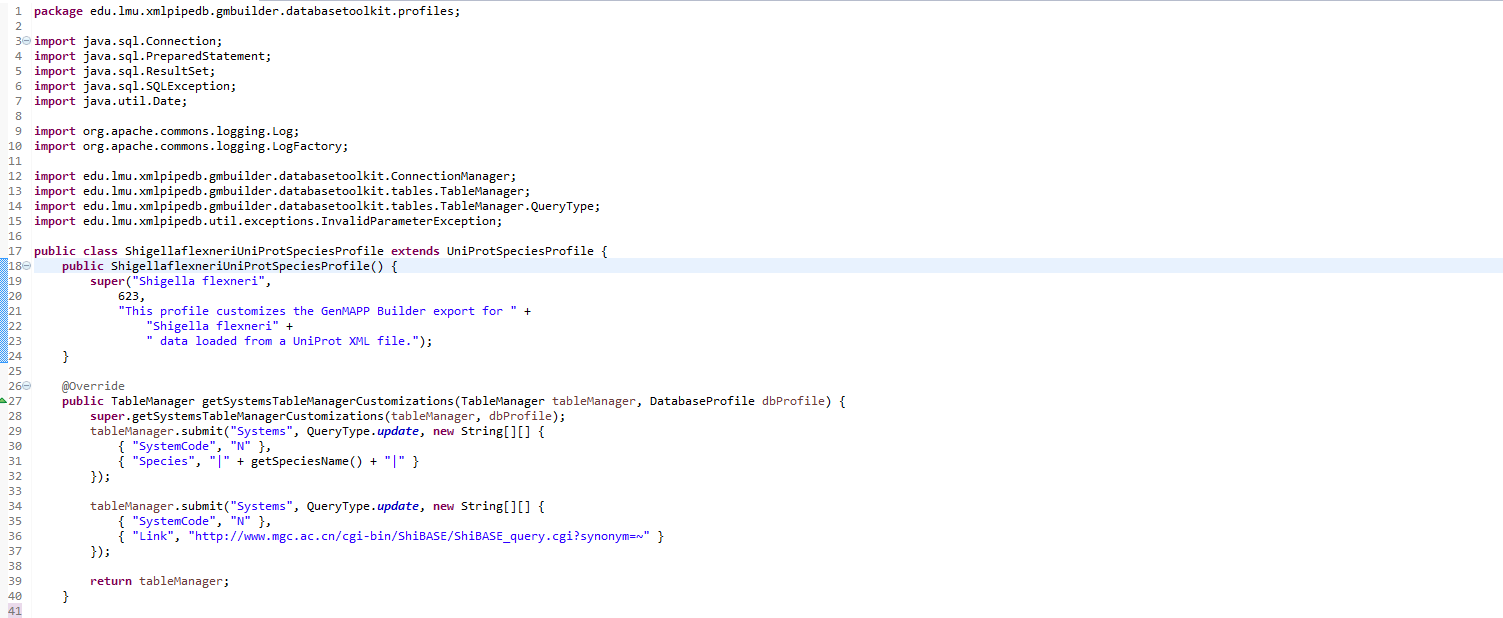
This is what the final customized class looks like:
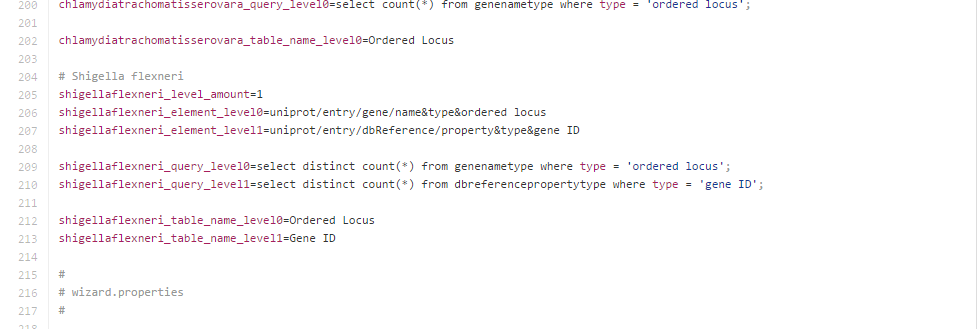
A further modification that was required was in the gmbuilder.properties file which we were supposed to edit to assist TallyEngine in its function, the initial customizations to this were the following lines:
This customization was insufficient in capturing the 92 missing genes. With the help of Dondi, Trixie and I replaced the insufficient SQL query with one that joined the missing 92 genes in the dbreference tag with the rest of the genes that were found by the default customization. Default customization can be found on Week 12 and Week 14. A sql union was used to execute this task which replaced the original sql query on gmbuilder.properties, as can be seen below:
select count(value) from (select value from genenametype where type = 'ordered locus' and value ~ '(CP|SF?)[0-9][0-9][0-9][0-9](\.[0-9])?' union select extra as value from (select propertytype.value as extra from propertytype inner join dbreferencetype on propertytype.dbreferencetype_property_hjid = dbreferencetype.hjid where dbreferencetype.type = 'EnsemblBacteria' and dbreferencetype.id ~ 'AAN[0-9][0-9][0-9][0-9][0-9]' and propertytype.type = 'gene ID' and propertytype.value ~ 'SF[0-9][0-9][0-9][0-9]') as f left join (select value from genenametype where type = 'ordered locus' and value ~ '(CP|SF?)[0-9][0-9][0-9][0-9](\.[0-9])?') as g on f.extra = g.value where g.value is null) as combined;
Individual Reflection
Statement of Work
- Describe exactly what you did on the project.
Three files, the XML Uniprot file, the GOA file, and the GO-OBO file were all imported into GenMAPP Builder by the QA. These files hold the information to create a gene database (.gdb), where I come in is this second step of exporting the files into a .gdb. I created a custom species profile for Shigella flexneri within the Java code of GenMAPP Builder. In order to do this I had to make my own distribution of GenMAPP Builder, and my own branch to commit to on GitHub. The edits I made to the code were the following:
I created a new subclass of UniProtSpeciesProfile called ShigellaflexneriUniprotSpeciesProfile. Within this new class I created a custom constructor that called the superclass's constructor and passed it information about Shigella flexneri, such as the taxonomy ID, and a custom String. I also overrode a method "getSystemsTableManagerCustomizations()" which, from my understanding, captures genes from the XML file we imported. The code for these initial customizations were provided to us by Dondi. However, as to be expected, the GenMAPP users alerted to us that we were missing 416 genes with these customizations. Trixie discovered that 92 of these genes could be found at unexpected locations in the XML file and so my task was to add another layer of customization. With help from Dondi, we overloaded the getSystemsTableManagerCustomizations method to account for these 92 genes in the XML file. Dondi added a SQL query within the method that captured the 92 missing genes and added them to the .gdb file. This was as many of the missing genes we could add. Throughout all of this I committed and pulled changes as needed.
Another customization that was required was within TallyEngine's gmbuilder.properties file. We added special properties for Shigella flexneri. These changes were pretty standard as they were in our class instructions, however, due to the nature of our GMBuilder java customizations, we need to make sure that TallyEngine could gather both the original genes and the 92 missing ones that were added on later. So, with help from Dondi, we added a SQL query that would be able to do this. The query used the union operation to get both the initial customization genes and the 92 missing genes.
These were the major changes I contributed to complete our project. Step by step can be seen on Week 14 and Week 15
- Provide references or links to artifacts of your work, such as:
- Wiki Pages
- Other files or documents
- Code or scripts
Assessment of Project
- Give an objective assessment of the success of your project workflow and teamwork.
- What worked and what didn't work?
- What would you do differently if you could do it all over again?
- Evaluate the Gene Database Project and Group Report in the following areas:
- Content: What is the quality of the work?
- Organization: Comment on the organization of the project and of your group's wiki pages.
- Completeness: Did your team achieve all of the project objectives? Why or why not?
Reflection on the Project
- What did you learn?
- With your head?((biological or computer science principles))
- With your heart?(personal qualities and teamwork qualities that make things work or not work)
- With your hands?(With your hands (technical skills))
- What lesson will you take away from this project that you will still use a year from now?
BIOL 367, Fall 2015, User Page, Team Page
| Weekly Assignments | Individual Journal Pages | Shared Journal Pages |
|---|---|---|
|
|
|
|