Anuvarsh Week 9
Contents
- 1 Export Information
- 2 TallyEngine
- 3 Using XMLPipeDB match to Validate the XML Results from the TallyEngine
- 4 Using SQL Queries to Validate the PostgreSQL Database Results from the TallyEngine
- 5 OriginalRowCounts Comparison
- 6 Visual Inspection
- 7 .gdb Use in GenMAPP
- 8 Compare Gene Database to Outside Resource
- 9 Other Links
Export Information
Version of GenMAPP Builder:
Computer on which export was run: back row, second from door.
Postgres Database name:
UniProt XML filename (give filename and upload and link to compressed file):
- UniProt XML version (The version information can be found at the UniProt News Page): UniProt release 2015_10
- UniProt XML download link: http://www.uniprot.org/uniprot/?query=organism:243277
- Time taken to import: 3.06 minutes
- Note:
GO OBO-XML filename (give filename and upload and link to compressed file):
- GO OBO-XML version (The version information can be found in the file properties after the file downloaded from the [GO Download page] has been unzipped): 10/27/2015, 2:24am
- GO OBO-XML download link: http://geneontology.org/page/download-ontology#Legacy_Downloads
- Time taken to import: 7.58 minutes
- Time taken to process: 4.37 minutes
- Note:
GOA filename (give filename and upload and link to compressed file):
- GOA version (News on this page records past releases; current information can be found in the Last modified field on the FTP site): 10/13/15
- GOA download link: http://ftp.ebi.ac.uk/pub/databases/GO/goa/proteomes/46.V_cholerae_ATCC_39315.goa
- Time taken to import:
- Note:
Name of .gdb file (give filename and upload and link to compressed file):
- Time taken to export: 0.06 minutes
- Start time: 10/27/2015, 3:52:08PM PDT
- End time: 10/27/2015, 5:09:55PM PDT
Note:
TallyEngine
- I ran the TallyEngine in GenMAPP Builder and recorded the number of records for UniProt and GO in the XML data and in the Postgres databases.
- After running PostgreSQL and making sure my database was running, I ran GenMAPP builder and connected it to the database.
- After performing an import, I chose Run XML and Database Tallies for Uniprot and and selected the UniProt and GO files that I imported.
- My Tally results are in the screenshot below:
-
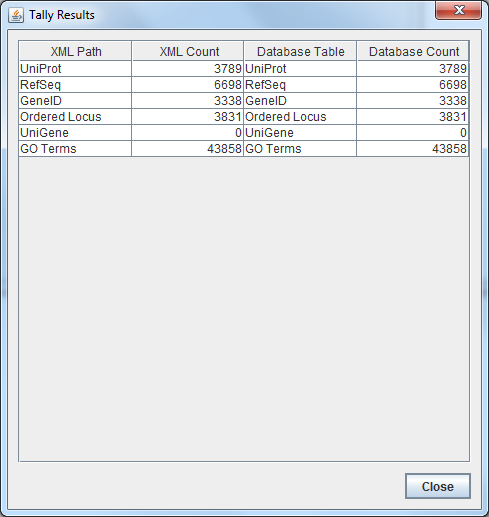
- The Tally results indicated counts for unique genes (labelled as Ordered Locus)
- XML: 3831 unique genes
- Database: 3831 unique genes
Using XMLPipeDB match to Validate the XML Results from the TallyEngine
- In order to check the number of genes via xmlpipedb match, I used a command that utilized match for the file containing all of the uniprot genes.
- Command Used:
java -jar xmlpipedb-match-1.1.1.jar "VC_[0-9][0-9][0-9][0-9]" < uniprot-organism%3A243277.xml"

- XMLPipeDB Match returned 2738 unique genes.
- This number is different than the numbers the TallyEngine returned.
- Upon later analysis (documented later in this lab notebook), I realized that the discrepancy between these counts is mostly attributed to the several different formats between which the genes are presented in the UniProt file. Despite all of these formats, we only counted the genes named in the format "VC_####" which disregards the other legitimate naming format.
- We modified our command to take into consideration both 'VC_####' and 'VC_A####' as legitimate gene names. This resulted in xmlpipedb-match indicating 3831 unique genes.
- These new results match the results of the Tally Engine.
Using SQL Queries to Validate the PostgreSQL Database Results from the TallyEngine
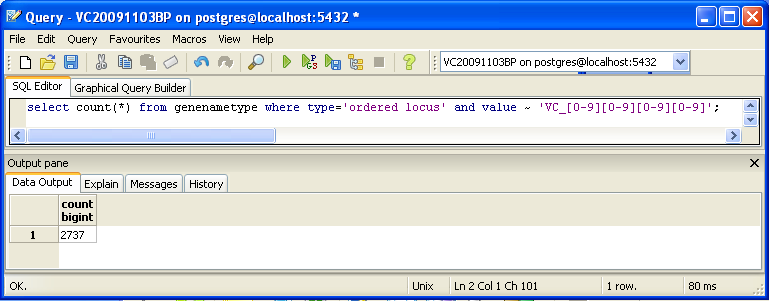
- On PostgreSQL, I searched through the database using a variation of the select count(*) query.
- Command used:
select count(*) from genenametype where type = 'ordered locus' and value ~ 'VC_[0-9][0-9][0-9][0-9]';
- PostgreSQL returned 2737 unique genes.
- These results are different for the same reason as the xmlpipedb-match results. This search only counted one format of gene names.
- When we reformatted the query to take into account both 'VC_####' and 'VC_A####', PostgreSQL indicated 3831 unique genes.
- This new count is the same as the xml and database results from the TallyEngine.
OriginalRowCounts Comparison
Within the .gdb file, look at the OriginalRowCounts table to see if the database has the expected tables with the expected number of records. Compare the tables and records with a benchmark .gdb file.
Benchmark .gdb file:
Copy the OriginalRowCounts table from the benchmark and new gdb and paste them here:
Note:
Visual Inspection
Perform visual inspection of individual tables to see if there are any problems.
- Look at the Systems table. Is there a date in the Date field for all gene ID systems present in the database?
- Open the UniProt, RefSeq, and OrderedLocusNames tables. Scroll down through the table. Do all of the IDs look like they take the correct form for that type of ID?
Note:
.gdb Use in GenMAPP
Note:
Putting a gene on the MAPP using the GeneFinder window
- Try a sample ID from each of the gene ID systems. Open the Backpage and see if all of the cross-referenced IDs that are supposed to be there are there.
Note:
Creating an Expression Dataset in the Expression Dataset Manager
- How many of the IDs were imported out of the total IDs in the microarray dataset? How many exceptions were there? Look in the EX.txt file and look at the error codes for the records that were not imported into the Expression Dataset. Do these represent IDs that were present in the UniProt XML, but were somehow not imported? or were they not present in the UniProt XML?
Note:
Coloring a MAPP with expression data
Note:
Running MAPPFinder
Note:
Compare Gene Database to Outside Resource
The OrderedLocusNames IDs in the exported Gene Database are derived from the UniProt XML. It is a good idea to check your list of OrderedLocusNames IDs to see how complete it is using the original source of the data (the sequencing organization, the MOD, etc.) Because UniProt is a protein database, it does not reference any non-protein genome features such as genes that code for functional RNAs, centromeres, telomeres, etc.
Note:
Other Links
User Page: Anindita Varshneya
Class Page: BIOL/CMSI 367: Biological Databases, Fall 2015
Group Page: GÉNialOMICS
Assignment Pages
Week 1 Assignment
Week 2 Assignment
Week 3 Assignment
Week 4 Assignment
Week 5 Assignment
Week 6 Assignment
Week 7 Assignment
Week 8 Assignment
Week 9 Assignment
Week 10 Assignment
Week 11 Assignment
Week 12 Assignment
No Week 13 Assignment
Week 14 Assignment
Week 15 Assignment
Individual Journals
Individual Journal Week 2
Individual Journal Week 3
Individual Journal Week 4
Individual Journal Week 5
Individual Journal Week 6
Individual Journal Week 7
Individual Journal Week 8
Individual Journal Week 9
Individual Journal Week 10
Individual Journal Week 11
Individual Journal Week 12
Individual Journal Week 14
Individual Journal Week 15
Class Journal Week 1
Class Journal Week 2
Class Journal Week 3
Class Journal Week 4
Class Journal Week 5
Class Journal Week 6
Class Journal Week 7
Class Journal Week 8
Class Journal Week 9
GÉNialOMICS Journal Week 10
GÉNialOMICS Journal Week 11
GÉNialOMICS Journal Week 12
GÉNialOMICS Journal Week 14
GÉNialOMICS Journal Week 15