Troque Week 9
Contents
Export Information
Version of GenMAPP Builder:
- gmbuilder-3.0.0-build-5
Computer on which export was run:
- The right-most computer at the very back of the room when facing the wide whiteboard/the computer at the back row that is closest to the door.
Postgres Database name:
- V_cholerae_20151027_gmb-5_TR
UniProt XML filename (give filename and upload and link to compressed file):
- UniProt XML version (The version information can be found at the UniProt News Page): UniProt release 2015_10
- UniProt XML download link: http://www.uniprot.org/uniprot/?query=organism:243277
- Time taken to import: 2.91 minutes
- Note:
GO OBO-XML filename (give filename and upload and link to compressed file):
- GO OBO-XML version (The version information can be found in the file properties after the file downloaded from the GO Download page has been unzipped): Version created on 10/27/2015 (at 2:24 AM)
- GO OBO-XML download link: http://geneontology.org/page/download-ontology#Legacy_Downloads
- Time taken to import: 6.93 minutes
- Time taken to process: 4.26 minutes
- Note:
GOA filename (give filename and upload and link to compressed file):
- GOA version (News on this page records past releases; current information can be found in the Last modified field on the FTP site): Version released on 10/14/15.
- GOA download link: http://ftp.ebi.ac.uk/pub/databases/GO/goa/proteomes/46.V_cholerae_ATCC_39315.goa
- Time taken to import: 0.06 minutes
- Note:
Name of .gdb file (give filename and upload and link to compressed file):
- Time taken to export: 5 hours, 33 minutes and 40 seconds
- Start time: 3:52:13 PM PDT
- End time: 9:25:53 PM PDT
- Note: I was out of town (went to D.C./Maryland for a research conference) so I couldn't check when the export finished, but it was a good thing that I logged in as myself (instead of the general student account) on the computer since it preserved the windows I had open from before I was left.
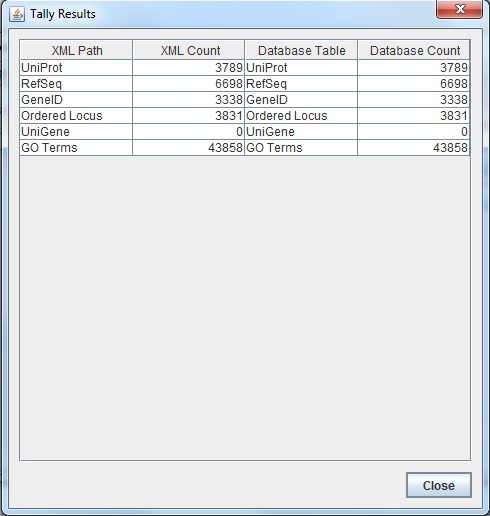
TallyEngine
- Run the TallyEngine in GenMAPP Builder and record the number of records for UniProt and GO in the XML data and in the Postgres databases.
- Choose the menu item Tallies > Run XML and Database Tallies for UniProt and GO...
- Choose the UniProt and GO OBO XML files that was uploaded from the previous sections of this assignment.
- Here is the screenshot of the tally result:
Using XMLPipeDB match to Validate the XML Results from the TallyEngine
Follow the instructions found on this page to run XMLPipeDB match.
- Note: When I tried downloading the jar file, it wasn't in the zip file that I downloaded when I clicked the link provided in the protocol. However, thanks to Kevin Wyllie who gave me the file, I managed to acquire it.
- Note: I created my own directory (called TrixRoq) in order to store all the files I downloaded into one single folder to differentiate it from the rest of the files in the ThawSpace disk. I also made a directory (called Working Data) in the ThawSpace0 disk in order to separate the source data from the data I worked with (per class suggestions from Dr. Dahlquist).
- As a result, I had to cd to these directories first before using the command for using Match.
- In order to change into the ThawSpace0\TrixRoq\Working Data directory, use the following commands on the command prompt window:
T: && cd "TrixRoq\Working Data"
- The command I used once inside the directory I want is:
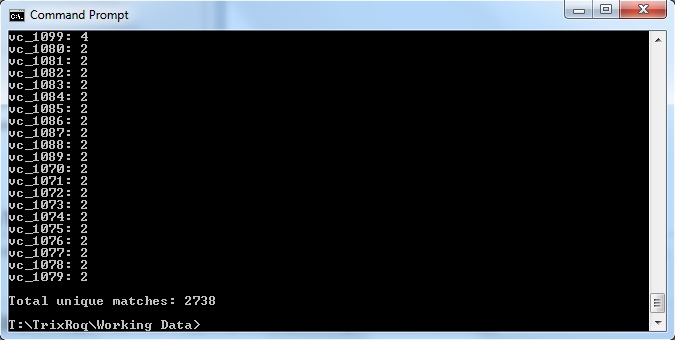
java -jar xmlpipedb-match-1.1.1.jar "VC_[0-9][0-9][0-9][0-9]" < uniprot-organism%3A243277.xml"
- The results are as follows:
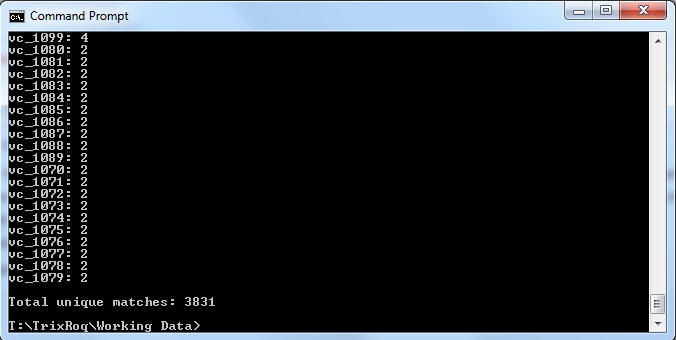
These results did not match up with what the TallyEngine gave (TallyEngine: 3831 vs. Match: 2738)
- As a result, the commands would have to be modified somehow so that the numbers match. Since the original command only accounted for the pattern with VC_####, we also have to account for the pattern for VC_A?####. The new command is as follows:
java -jar xmlpipedb-match-1.1.1.jar "VC_A?[0-9][0-9][0-9][0-9]" < uniprot-organism%3A243277.xml > command_prompt_results_TR.txt
which gave the results:
and saved it to the file "command_prompt_results_TR.txt"
Using SQL Queries to Validate the PostgreSQL Database Results from the TallyEngine
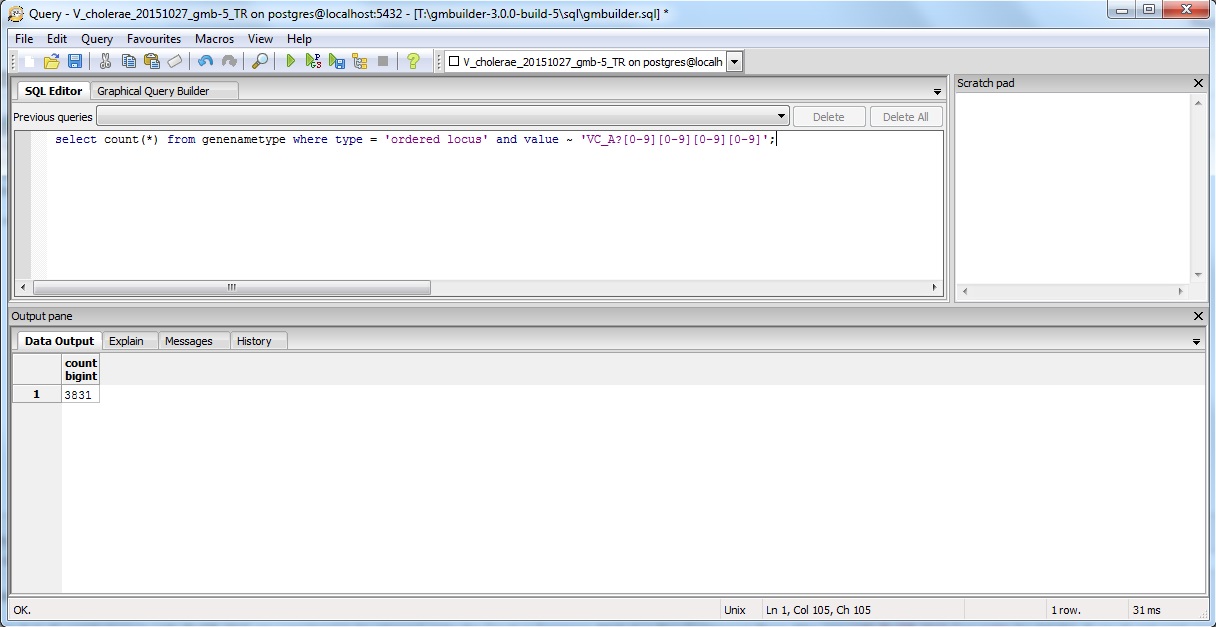
Since I already know to look for the pattern VC_A?#### instead of just VC_####, I would type in the following command in the pgAdmin window:
select count(*) from genenametype where type = 'ordered locus' and value ~ 'VC_A?[0-9][0-9][0-9][0-9]';
The results are as follows:
The result is the same for both TallyEngine and Match (i.e., 3831 counts).
OriginalRowCounts Comparison
Within the .gdb file, look at the OriginalRowCounts table to see if the database has the expected tables with the expected number of records. Compare the tables and records with a benchmark .gdb file.
Benchmark .gdb file:
Copy the OriginalRowCounts table from the benchmark and new gdb and paste them here:
Note:
Visual Inspection
Perform visual inspection of individual tables to see if there are any problems.
- Look at the Systems table. Is there a date in the Date field for all gene ID systems present in the database?
- Open the UniProt, RefSeq, and OrderedLocusNames tables. Scroll down through the table. Do all of the IDs look like they take the correct form for that type of ID?
Note:
.gdb Use in GenMAPP
Note:
Putting a gene on the MAPP using the GeneFinder window
- Try a sample ID from each of the gene ID systems. Open the Backpage and see if all of the cross-referenced IDs that are supposed to be there are there.
Note:
Creating an Expression Dataset in the Expression Dataset Manager
- How many of the IDs were imported out of the total IDs in the microarray dataset? How many exceptions were there? Look in the EX.txt file and look at the error codes for the records that were not imported into the Expression Dataset. Do these represent IDs that were present in the UniProt XML, but were somehow not imported? or were they not present in the UniProt XML?
Note:
Coloring a MAPP with expression data
Note:
Running MAPPFinder
Note:
Assignment Links
Weekly Assignments
- Week 1
- Week 2
- Week 3
- Week 4
- Week 5
- Week 6
- Week 7
- Week 8
- Week 9
- Week 10
- Week 11
- Week 12
- No Week 13 Assignment
- Week 14
- Week 15
Individual Journal Entries
- Week 1 - This is technically the user page.
- Week 2
- Week 3
- Week 4
- Week 5
- Week 6
- Week 7
- Week 8
- Week 9
- Week 10
- Week 11
- Week 12
- No Week 13 Assignment
- Week 14
- Week 15