Gene Database Testing Report- cw20151203
Contents
- 1 Files Asked for in the Gene Database Testing Report
- 2 Pre-requisites
- 3 Gene Database Creation
- 4 Gene Database Testing Report
- 4.1 Export Information
- 4.2 TallyEngine
- 4.3 Using XMLPipeDB match to Validate the XML Results from the TallyEngine
- 4.4 Using SQL Queries to Validate the PostgreSQL Database Results from the TallyEngine
- 4.5 OriginalRowCounts Comparison
- 4.6 Visual Inspection
- 4.7 .gdb Use in GenMAPP
- 4.8 Compare Gene Database to Outside Resource
Files Asked for in the Gene Database Testing Report
For convenience, all of the files explicitly asked for in the sections below were compressed together in this file: [[]]
Pre-requisites
The following set of software was used in the creation and testing of the Bordetella pertussis gene database:
- 7-ziptool that for unpacking .gz and .zip files
- PostgreSQL on Windows (version 9.4.x)
- GenMAPP Builder
- Java JDK 1.8 64-bit
- GenMAPP 2
- XMLPipeDB match utility for counting IDs in XML files
- Microsoft Access for reading .mdb files
Gene Database Creation
Downloading Data Source Files and GenMAPP Builder
- I download the UniProt XML, GOA, and GO OBO-XML files for Bordetella Pertussis along with the GenMAPP Builder program.
- All files were saved to the folder Bklein7_CW\bpertussis_cw20151203 on my computer's ThawSpace.
- Files that required extraction were unzipped using 7-zip.
- Data files that remained in a folder after unzipping were removed from their folders to facilitate organization and command line processing.
UniProt XML
- I went to the UniProt Complete Proteomes page.
- From there, I navigated to the complete proteome download page for Bordetella pertussis (strain Tohama I / ATCC BAA-589 / NCTC 13251).
- I clicked on the "Download" button at the top of the page above and selected the following options:
- "Download all"
- "XML" from the "Format" drop-down menu
- "Compressed" format
- I extracted the file using 7-zip.
GOA
- UniProt-GOA files can be downloaded from the UniProt-GOA ftp site.
- Within the above site, I navigated to the for Bordetella pertussis strain Tohama I.
- This text file was automatically opened by my browser. Therefore, I had to manually download the file.
GO OBO-XML
- I downloaded the GO OBO-XML formatted file from the Gene Ontology legacy download page.
- I extracted the file using 7-zip.
Downloaded GenMAPP Builder
- I downloaded the custom version of GenMAPP Builder including the Bordetella pertussis custom class expanded to include ORF listings in exports (Version 3.0.0 Build 5 - cw20151203): File:Dist cw20151203.zip.
- I extracted the GenMAPP Builder folder using 7-zip.
Creating the New Database in PostgreSQL
- I launched pgAdmin III and connected to the PostgreSQL 9.4 server (localhost:5432).
- On this server, I created a new database: bpertussis_cw20151201_gmb3build5.
- I opened the SQL Editor tab to use an XMLPipeDB query to create the tables in the database.
- I clicked on the Open File icon and selected the file gmbuilder.sql. This imported a series of SQL commands into the editor tab.
- I clicked on the Execute Query icon to run this command.
- In viewing the schema for this database, I confirmed that there were 167 tables after running the above command.
Configuring GenMAPP Builder to Connect to the PostgreSQL Database
- To begin, I launched gmbuilder.bat.
- I selected the "Configure Database" option and entered the following information into the fields below:
- Host or address: localhost
- Port number: 5432
- Database name: bpertussis_cw20151201_gmb3build5
- Username: postgres
- Password: Welcome1
Importing Data into the PostgreSQL Database
- The downloaded data files for Bordetella pertussis were specified and imported into the database by clicking on the following buttons:
- Selected File > Import UniProt XML...
- Selected File > Import GO OBO-XML...
- Clicked OK to the message asking to process the GO data.
- Selected File > Import GOA...
Exporting a GenMAPP Gene Database (.gdb)
- I selected File > Export to GenMAPP Gene Database... to begin the export process.
- I typed my name in the owner field (Brandon Klein).
- I selected "Bordetella pertussis (strain Tohama I/ATCC BAA-589/NCTC 13251), Taxon ID 257313" as the gene database species and then clicked Next.
- The database was saved as bpertussis-std_cw20151203.
- I checked the boxes for exporting all Molecular Function, Cellular Component, and Biological Process Gene Ontology Terms.
- Finally, I clicked the "Next" button to begin the export process.
Gene Database Testing Report
Export Information
Version of GenMAPP Builder: Version 3.0.0 Build 5 - cw20151203
Computer on which export was run: Seaver 120- Last computer on the right in the row farthest from the front of the room
Postgres Database name: bpertussis_cw20151201_gmb3build5
UniProt XML filename: File:Uniprot-proteome-UP000002676 cw20151201.zip
- UniProt XML version (The version information was found at the UniProt News Page): 2015_11
- UniProt XML download link: Bordetella pertussis (strain Tohama I / ATCC BAA-589 / NCTC 13251)
- Time taken to import: 2.59 minutes
- Note: The import time was nearly equivalent to that when creating the previous "Bordetella pertussis" gene database: bpertussis-std_cw20151119.gdb (2.60 minute). No interruptions occurred during this process.
GO OBO-XML filename: File:Go daily-termdb cw20151201.zip
- GO OBO-XML version (The version information was found in the file properties): Last Modified- December 01, 2015, 2:21:31 AM
- GO OBO-XML download link: Gene Ontology legacy download page
- Time taken to import: 7.08 minutes
- Time taken to process: 4.42 minutes
- Note: The import and processing times were similar to those for the previous "Bordetella pertussis" gene database: bpertussis-std_cw20151119.gdb (6.99 minutes and 4.48 minutes respectively). No interruptions occurred during these processes.
GOA filename: File:145.B pertussis ATCC BAA-589 cw20151201.zip
- GOA version (found in the Last modified field on the FTP site): Last Modified- 11/10/15 1:39:00 PM
- GOA download link: for Bordetella pertussis strain Tohama I
- Time taken to import: 0.04 minutes
- Note: The import time was equal to that of the previous "Bordetella pertussis" gene database: bpertussis-std_cw20151119.gdb. No interruptions occurred during this process.
Name of .gdb file: File:Bpertussis-std cw20151203.zip
- Time taken to export:
- Start time: 4:02 PM
- End time: 4:56 PM
- Elapsed time: 54 minutes
Note: No interruptions occurred during the export process.
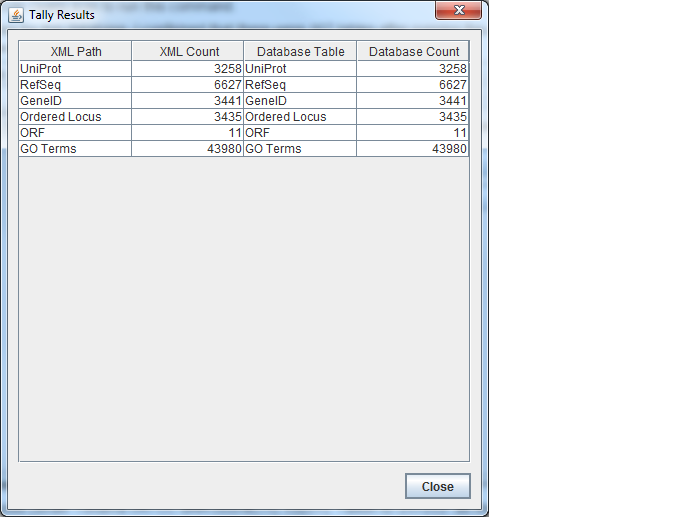
TallyEngine
- I ran the TallyEngine in GenMAPP Builder and specified the following files:
- Results:
Using XMLPipeDB match to Validate the XML Results from the TallyEngine
The following functions were performed using the Windows command line (cmd).
- I entered my project folder using the following command:
cd /d T:\Bklein7_CW
- I used XMLPipeDB match to identify matches of any ordered locus name following the pattern "BP####" in the UniProt XML file. The command sequence used is as follows:\
java -jar xmlpipedb-match-1.1.1.jar "BP[0-9][0-9][0-9][0-9]" < "uniprot-proteome%3AUP000002676_cw20151201.xml"
- Match Results:
- make note to ask dondi specific code to use to obtain 3446 match results
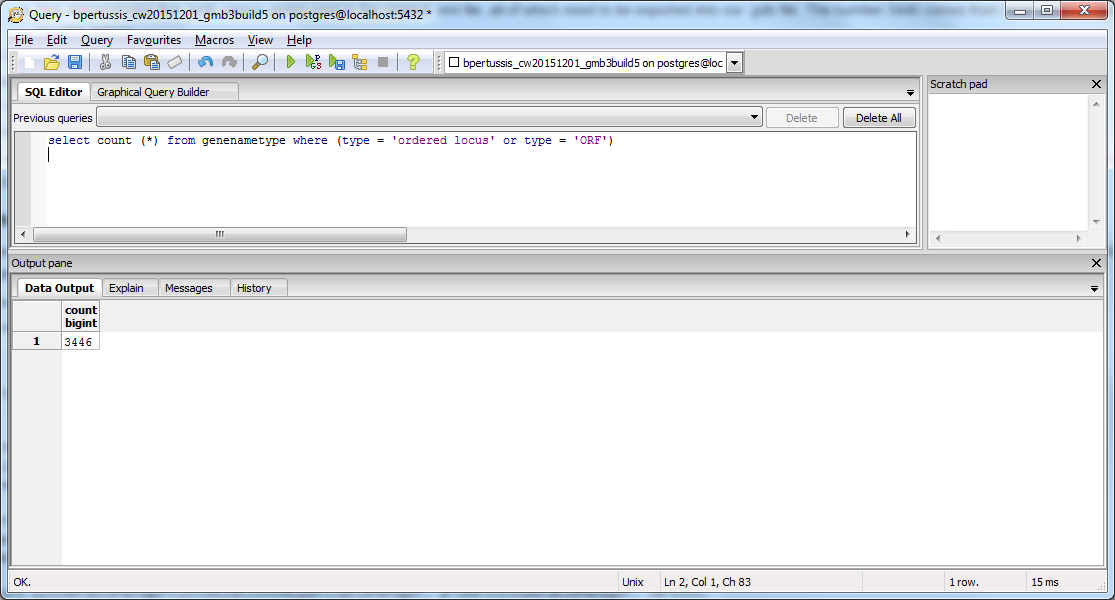
Using SQL Queries to Validate the PostgreSQL Database Results from the TallyEngine
I ran a SQL query designed to match the pattern BP####:
select count (*) from genenametype where type = 'ordered locus' and value ~ 'BP[0-9][0-9][0-9][0-9]';
Results:
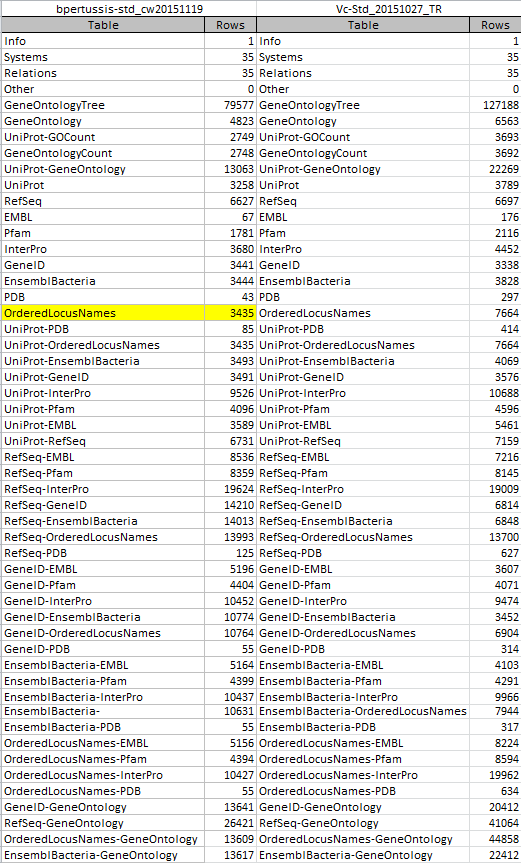
OriginalRowCounts Comparison
I opened the gene database file File:Bpertussis-std cw20151203.zip in Microsoft Access and assessed the "OriginalRowCounts" table to see if the expected tables were listed with the expected number of records. The contents of this table were compared to the OriginalRowCounts table of an existing .gdb file created during Week 9.
Benchmark .gdb file: Vc-Std_20151027_TR
"OriginalRowCounts" table from the benchmark and new gdb:
- All 52 tables present in the 2015 Vibrio cholerae database were also present in the B. pertussis database tagged _cw20151203. This confirmed that all expected tables were successfully created.
- Further, the "OrderedLocusNames" count produced by Tally Engine should hopefully represent this number.
Note: The "OriginalRowCounts" tables were too large to screenshot. To circumvent this problem and facilitate the comparison, I copied the "OriginalRowCounts" tables from both gene databases into an Excel file and zoomed out. The above screenshot was taken from this Excel file. The "OrderedLocusNames" row counts are highlighted in yellow.
Visual Inspection
I visually inspected individual tables within File:Bpertussis-std cw20151203.zip using Microsoft Access.
- Systems Table
- 35 gene ID systems were listed, 11 of which listed the appropriate import date (12/03/2015)
- All gene ID systems relevant to B. pertussis were listed. This includes: EMBL, EnsemblBacteria, GeneID, GeneOntology, InterPro, OrderedLocusNames, Pfam, RefSeq, and UniProt.
- This result corresponded with that of the benchmark .gdb file listed in the "OriginalRowCounts Comparison" section.
- 35 gene ID systems were listed, 11 of which listed the appropriate import date (12/03/2015)
- UniProt Table
- RefSeq Table
- This table contained 6627 entries. All IDs began with one of three prefixes: "NP_", "YP_", or "WP_". The meanings of these prefixes can be found in the RefSeq documentation found here.
- "NP_" and "YP_" Prefixes
- Refer to proteins. There are 3410 "NP_" IDs and 7 "YP_" IDs.
- "WP_" Prefixes
- Refer to "autonomous non-redundant proteins that are not yet directly annotated on a genome". There were 3210 IDs with the "WP_" prefixes.
- Overall, every entry in the ID column was an expected value.
- "NP_" and "YP_" Prefixes
- This table contained 6627 entries. All IDs began with one of three prefixes: "NP_", "YP_", or "WP_". The meanings of these prefixes can be found in the RefSeq documentation found here.
- OrderedLocusNames Table
- This table contained 3446 entries (consistent with & SQL counts).
- The IDs were copied into an Excel document for analysis:
- 3434 IDs conformed to the pattern "BP####"
- 1 ID was unique: "BP3167.1"
- The other 11 IDs showed up as ORF
.gdb Use in GenMAPP
- By following the instructions in Part 2 of the Vibrio cholerae Microarray Data Analysis and looking at Brandon's Week 9 individual journal assignment, I was able to verify that our Gene Database works in GenMAPP.
- I was able to open the GenMAPP program on the computer, and then I went to Data -> Choose Gene Database -> and selected the cw20151119 gdb file.
- There were no problems thus far as our database was able to load into the program.
Putting a gene on the MAPP using the GeneFinder window
- In the main GenMAPP Drafting Board window, I left-clicked on the icon for "Gene" in the upper left corner of the window.
- I clicked on the Drafting Board to place the Gene on the MAPP.
- Then I right-clicked on the gene to access the GeneFinder window.
- I went through each of the five inconsistent Gene IDs in GeneFinder.
- I typed in the gene Id BP3167.1 into the Gene ID field and selected "OrderedLocusName" for the Gene ID system.
- In the main GenMAPP Drafting Board window, I left-clicked on the icon for "Gene" in the upper left corner of the window.
- I clicked on the Drafting Board to place the Gene on the MAPP.
- Then I right-clicked on the gene to access the GeneFinder window.
- I typed in the gene Id BP1252 into the Gene ID field and selected "OrderedLocusName" for the Gene ID system. This served as a "control" to look up a consistent Gene ID to compare to.
- In the main GenMAPP Drafting Board window, I left-clicked on the icon for "Gene" in the upper left corner of the window.
- I clicked on the Drafting Board to place the Gene on the MAPP.
- Then I right-clicked on the gene to access the GeneFinder window.
- In the main GenMAPP Drafting Board window, I left-clicked on the icon for "Gene" in the upper left corner of the window.
- I typed in the gene Id BP0101A into the Gene ID field and selected "OrderedLocusName" for the Gene ID system.
- In the main GenMAPP Drafting Board window, I left-clicked on the icon for "Gene" in the upper left corner of the window.
- I clicked on the Drafting Board to place the Gene on the MAPP.
- Then I right-clicked on the gene to access the GeneFinder window.
- In the main GenMAPP Drafting Board window, I left-clicked on the icon for "Gene" in the upper left corner of the window.
- I clicked on the Drafting Board to place the Gene on the MAPP.
- Then I right-clicked on the gene to access the GeneFinder window.
- I typed in the gene Id BP0101B into the Gene ID field and selected "OrderedLocusName" for the Gene ID system.
- In the main GenMAPP Drafting Board window, I left-clicked on the icon for "Gene" in the upper left corner of the window.
- I clicked on the Drafting Board to place the Gene on the MAPP.
- Then I right-clicked on the gene to access the GeneFinder window.
- I typed in the gene Id BP0684A into the Gene ID field and selected "OrderedLocusName" for the Gene ID system.
- When the gene was found this is the back page that popped up:
- In the main GenMAPP Drafting Board window, I left-clicked on the icon for "Gene" in the upper left corner of the window.
- I clicked on the Drafting Board to place the Gene on the MAPP.
- Then I right-clicked on the gene to access the GeneFinder window.
- I typed in the gene Id BP0970A into the Gene ID field and selected "GeneID" for the Gene ID system.
- All of the expected cross-referenced IDs were present.
- Screenshot of all of the sample ID's on a MAPP:
- Genesonmap_cw20151203.png
Expression Dataset and MAPPFinder Analysis
- We do not have the expression dataset yet that is to be created by the GenMAPP Builders; they are still working on performing the corrections to the data that has been compiled into an excel spreadsheet.
- Once the file is complete, we will proceed with the data analysis using the desired programs.
Compare Gene Database to Outside Resource
- We will complete this step after progressing further into the project.
The OrderedLocusNames IDs in the exported Gene Database are derived from the UniProt XML. It is a good idea to check your list of OrderedLocusNames IDs to see how complete it is using the original source of the data (the sequencing organization, the MOD, etc.) Because UniProt is a protein database, it does not reference any non-protein genome features such as genes that code for functional RNAs, centromeres, telomeres, etc.