Difference between revisions of "Aporras1 Electronic Notebook"
From LMU BioDB 2017
(RNA 1 reading frames) |
(→Study the curl'ed Code: added numbering and grep/sed) |
||
| (30 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | 5'-cgtatgctaataccatgttccgcgtataacccagccgccagttccgctggcggcatttta-3' Template DNA strand ( | + | =Week 2= |
| − | 3'-gcatacgattatggtacaaggcgcatattgggtcggcggtcaaggcgaccgccgtaaaat-5' Complimentary DNA Strand ( | + | 5'-cgtatgctaataccatgttccgcgtataacccagccgccagttccgctggcggcatttta-3' Template DNA strand (Top Strand) |
| + | 3'-gcatacgattatggtacaaggcgcatattgggtcggcggtcaaggcgaccgccgtaaaat-5' Complimentary DNA Strand (Bottom Strand) | ||
| − | 3'-gcauacgauuaugguacaaggcgcauauugggucggcggucaaggcgaccgccguaaaau-5' RNA Strand ( | + | 3'-gcauacgauuaugguacaaggcgcauauugggucggcggucaaggcgaccgccguaaaau-5' RNA Strand (Top Strand) |
| − | 5'-cguaugcuaauaccauguuccgcguauaacccagccgccaguuccgcuggcggcauuuua-3' RNA Strand ( | + | 5'-cguaugcuaauaccauguuccgcguauaacccagccgccaguuccgcuggcggcauuuua-3' RNA Strand (Bottom Strand) |
*Completed letter by letter strand of the complimentary DNA using the template strand | *Completed letter by letter strand of the complimentary DNA using the template strand | ||
| Line 10: | Line 11: | ||
**changed T to U on the RNA strands | **changed T to U on the RNA strands | ||
| − | ==RNA Strand ( | + | ==RNA Strand (Top Strand) Reading Frames== |
*Put 1 single space every 3 codons starting after a, after c, and after g to create the 3 different reading frames | *Put 1 single space every 3 codons starting after a, after c, and after g to create the 3 different reading frames | ||
| − | + | (-1) 3'-gca uac gau uau ggu aca agg cgc aua uug ggu cgg cgg uca agg cga ccg ccg uaa aau-5' | |
| − | + | A-Y-D-Y-G-T-R-R-I-L-G-R-R-S-R-R-P-P-STOP-N | |
| − | + | ||
| + | (-2) 3'-g cau acg auu aug gua caa ggc gca uau ugg guc ggc ggu caa ggc gac cgc cgu aaa au-5' | ||
| + | g-H-T-I-M-V-Q-G-A-Y-W-V-G-G-Q-G-D-R-R-K-au | ||
| + | |||
| + | (-3) 3'-gc aua cga uua ugg uac aag gcg cau auu ggg ucg gcg guc aag gcg acc gcc gua aaa u-5' | ||
| + | gc-I-R-L-W-Y-K-A-H-I-G-S-A-V-K-A-T-A-V-K-u | ||
| + | |||
| + | ==RNA Strand (Bottom Strand)== | ||
| + | |||
| + | *Put 1 single space every 3 codons starting after a, after c, and after g to create the 3 different reading frames | ||
| + | |||
| + | (+1) 5'-cgu aug cua aua cca ugu ucc gcg uau aac cca gcc gcc agu ucc gcu ggc ggc auu uua-3' | ||
| + | R-M-L-I-P-C-S-A-Y-N-P-A-A-S-S-A-G-G-I-L | ||
| + | |||
| + | (+2) 5'-c gua ugc uaa uac cau guu ccg cgu aua acc cag ccg cca guu ccg cug gcg gca uuu ua-3' | ||
| + | c-V-C-STOP-Y-H-V-O-R-I-T-Q-P-P-V-P-L-A-A-F-ua | ||
| + | |||
| + | (+3) 5'-cg uau gcu aau acc aug uuc cgc gua uaa ccc agc cgc cag uuc cgc ugg cgg cau uuu a-3' | ||
| + | cg-Y-A-N-T-M-F-R-V-STOP-P-S-R-Q-F-R-W-R-H-F-a | ||
| + | |||
| + | =Week 3= | ||
| + | |||
| + | For the week 3 I chose the front page of [[http://www.foxnews.com Fox News]] to "hack" and change the source code. | ||
| + | |||
| + | '''Used "developer tools" in Google Chrome | ||
| + | |||
| + | #I changed 3 links underneath the main header. | ||
| + | #Uploaded an image of a teddy bear hamster to the LMU BioDB 2017. | ||
| + | #Changed the src code to the image of the hamster. | ||
| + | #Changed the header. | ||
| + | #Changed the header within the image. | ||
| + | |||
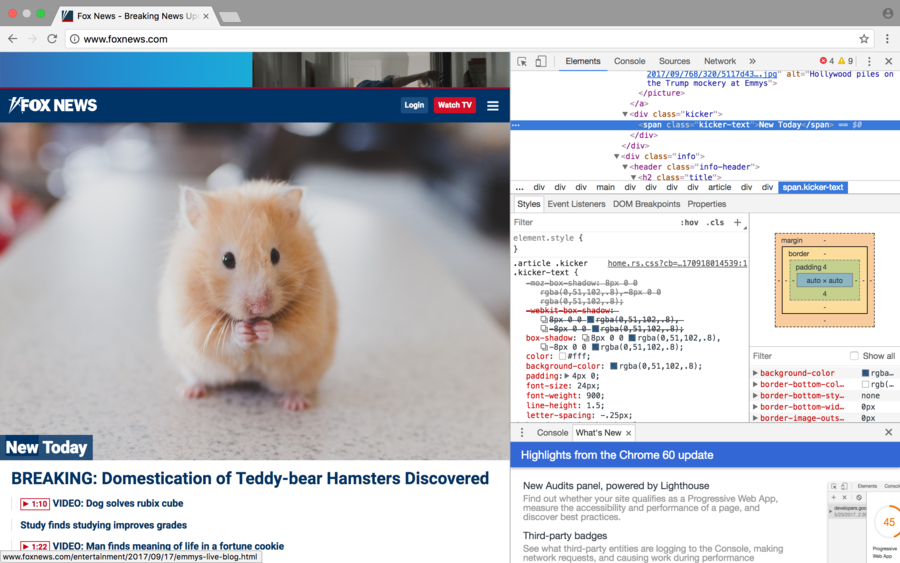
| + | =="Hacked" Page with Developer Tools== | ||
| + | |||
| + | [[File:Aporras1' Hacked Page with Developer Tools.png|900px|File:900pixels]] | ||
| + | |||
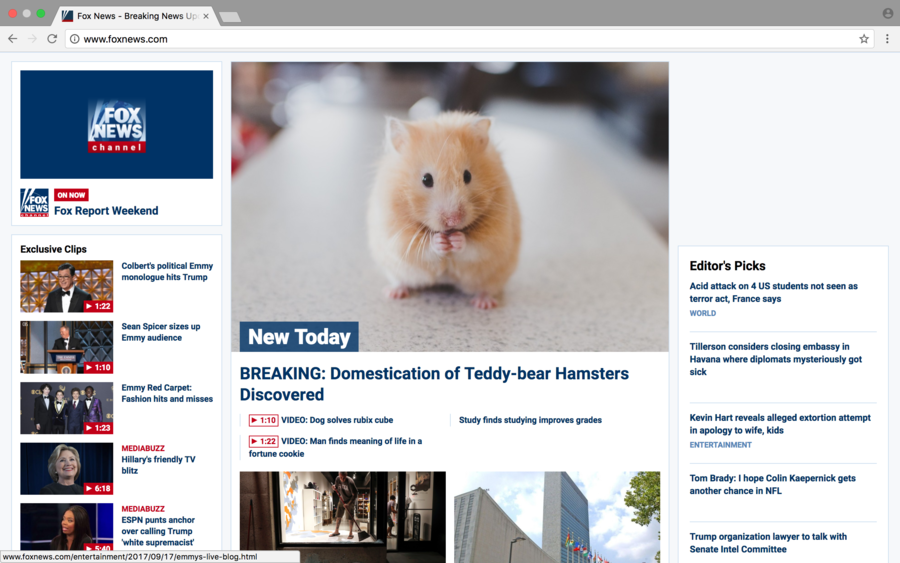
| + | =="Hacked" Page without Developer Tools== | ||
| + | |||
| + | [[File:Aporras1's_Hacked_Page_without_Developer_Tools.png|900px|File:900pixels]] | ||
| + | |||
| + | ==DMing the Server with curl== | ||
| + | |||
| + | #Used curl command in terminal and found the line when inspecting the text box: <form method=POST action="/cgi-bin/translate/dna_aa"> | ||
| + | #Inspected the button which resulted in translating the sequence: <input type="submit" value="TRANSLATE SEQUENCE"> | ||
| + | #Inspected the text box: <textarea name="pre_text" rows="15" cols="70"></textarea> | ||
| + | #Inferring from the code mentioned prior, the text within the box is known as "pre_text". | ||
| + | #Looked at various curl codes and found curl -d sends data as a POST as if the user has filled out certain forms. The examples used: -d name=Antonio if I was putting my name into a form. | ||
| + | #First attempt: curl -d "pre_text=cgtatgctaataccatgttccgcgtataacccagccgccagttccgctggcggcatttta" which did not work. | ||
| + | #Second attempt: curl -d "pre_text=cgtatgctaataccatgttccgcgtataacccagccgccagttccgctggcggcatttta" https://expasy.org/translate | ||
| + | #Consulted another source to realize I needed to also add &Submit=TRANSLATE SEQUENCE to the code along with the correct POST which would be "/cgi-bin/translate/dna_aa" | ||
| + | #Third attempt: curl -d "pre_text=cgtatgctaataccatgttccgcgtataacccagccgccagttccgctggcggcatttta&Submit=TRANSLATE SEQUENCE" https://expasy.org/cgi-bin/translate/dna_aa which also failed. | ||
| + | #Talked more with Simon and realized it should be "http://web.expasy.org/cgi-bin/translate/dna_aa" and not "https://expasy.org/cgi-bin/translate/dna_aa" | ||
| + | #Attempt 4: curl -d "pre_text=cgtatgctaataccatgttccgcgtataacccagccgccagttccgctggcggcatttta&Submit=TRANSLATE SEQUENCE" http://web.expasy.org/cgi-bin/translate/dna_aa | ||
| + | |||
| + | ==Study the curl'ed Code== | ||
| + | |||
| + | Below, going line by line clicking each link and describing where it leads | ||
| + | |||
| + | #http://www.w3.org/TR/html4/loose.dtd : Described as "the HTML 4.01 Transitional DTD, which includes presentation attributes and elements that W3C expects to phase out as support for style sheets matures." | ||
| + | #http://web.expasy.org/favicon.ico : The logo for Expasy. | ||
| + | #http://www.sib.swiss : The main web page for Swiss Institute of Bioinformatics. | ||
| + | #https://www.expasy.org : The Expasy Bioinformatics Resource Portal. | ||
| + | #http://web.expasy.org/translate/ : Expasy translation tool. | ||
| + | #https://en.wikipedia.org/wiki/Open_reading_frame : Wikipedia link to what an "open reading frame" is described as. | ||
| + | #https://www.expasy.org/disclaimer.html : Expasy disclaimer. | ||
| + | |||
| + | Identifiers: | ||
| + | #sib_top: The top of the webpage. | ||
| + | #sib_container: The "container" of the webpage. | ||
| + | #sib_header_small: The small header for the webpage. | ||
| + | #sib_body: The "body" of the webpage outside of the container. | ||
| + | #sib_footer: The footer at the bottom of the webpage. | ||
| + | #sib_expasy_logo: Logo at the top right of the page. | ||
| + | #resource_header: Header of the webpage. | ||
| + | #sib_header_nav: Homepage link at the top right of the page for navigation. | ||
| + | |||
| + | #Couldn't figure out how to use the grep and sed commands in command line. | ||
Latest revision as of 06:56, 20 September 2017
Contents
Week 2
5'-cgtatgctaataccatgttccgcgtataacccagccgccagttccgctggcggcatttta-3' Template DNA strand (Top Strand) 3'-gcatacgattatggtacaaggcgcatattgggtcggcggtcaaggcgaccgccgtaaaat-5' Complimentary DNA Strand (Bottom Strand)
3'-gcauacgauuaugguacaaggcgcauauugggucggcggucaaggcgaccgccguaaaau-5' RNA Strand (Top Strand)
5'-cguaugcuaauaccauguuccgcguauaacccagccgccaguuccgcuggcggcauuuua-3' RNA Strand (Bottom Strand)
- Completed letter by letter strand of the complimentary DNA using the template strand
- Made two RNA strands using the 5' to 3' DNA strand and the 3' to 5' DNA strand
- changed T to U on the RNA strands
RNA Strand (Top Strand) Reading Frames
- Put 1 single space every 3 codons starting after a, after c, and after g to create the 3 different reading frames
(-1) 3'-gca uac gau uau ggu aca agg cgc aua uug ggu cgg cgg uca agg cga ccg ccg uaa aau-5' A-Y-D-Y-G-T-R-R-I-L-G-R-R-S-R-R-P-P-STOP-N
(-2) 3'-g cau acg auu aug gua caa ggc gca uau ugg guc ggc ggu caa ggc gac cgc cgu aaa au-5' g-H-T-I-M-V-Q-G-A-Y-W-V-G-G-Q-G-D-R-R-K-au
(-3) 3'-gc aua cga uua ugg uac aag gcg cau auu ggg ucg gcg guc aag gcg acc gcc gua aaa u-5' gc-I-R-L-W-Y-K-A-H-I-G-S-A-V-K-A-T-A-V-K-u
RNA Strand (Bottom Strand)
- Put 1 single space every 3 codons starting after a, after c, and after g to create the 3 different reading frames
(+1) 5'-cgu aug cua aua cca ugu ucc gcg uau aac cca gcc gcc agu ucc gcu ggc ggc auu uua-3' R-M-L-I-P-C-S-A-Y-N-P-A-A-S-S-A-G-G-I-L
(+2) 5'-c gua ugc uaa uac cau guu ccg cgu aua acc cag ccg cca guu ccg cug gcg gca uuu ua-3' c-V-C-STOP-Y-H-V-O-R-I-T-Q-P-P-V-P-L-A-A-F-ua
(+3) 5'-cg uau gcu aau acc aug uuc cgc gua uaa ccc agc cgc cag uuc cgc ugg cgg cau uuu a-3' cg-Y-A-N-T-M-F-R-V-STOP-P-S-R-Q-F-R-W-R-H-F-a
Week 3
For the week 3 I chose the front page of [Fox News] to "hack" and change the source code.
Used "developer tools" in Google Chrome
- I changed 3 links underneath the main header.
- Uploaded an image of a teddy bear hamster to the LMU BioDB 2017.
- Changed the src code to the image of the hamster.
- Changed the header.
- Changed the header within the image.
"Hacked" Page with Developer Tools
"Hacked" Page without Developer Tools
DMing the Server with curl
- Used curl command in terminal and found the line when inspecting the text box: <form method=POST action="/cgi-bin/translate/dna_aa">
- Inspected the button which resulted in translating the sequence: <input type="submit" value="TRANSLATE SEQUENCE">
- Inspected the text box: <textarea name="pre_text" rows="15" cols="70"></textarea>
- Inferring from the code mentioned prior, the text within the box is known as "pre_text".
- Looked at various curl codes and found curl -d sends data as a POST as if the user has filled out certain forms. The examples used: -d name=Antonio if I was putting my name into a form.
- First attempt: curl -d "pre_text=cgtatgctaataccatgttccgcgtataacccagccgccagttccgctggcggcatttta" which did not work.
- Second attempt: curl -d "pre_text=cgtatgctaataccatgttccgcgtataacccagccgccagttccgctggcggcatttta" https://expasy.org/translate
- Consulted another source to realize I needed to also add &Submit=TRANSLATE SEQUENCE to the code along with the correct POST which would be "/cgi-bin/translate/dna_aa"
- Third attempt: curl -d "pre_text=cgtatgctaataccatgttccgcgtataacccagccgccagttccgctggcggcatttta&Submit=TRANSLATE SEQUENCE" https://expasy.org/cgi-bin/translate/dna_aa which also failed.
- Talked more with Simon and realized it should be "http://web.expasy.org/cgi-bin/translate/dna_aa" and not "https://expasy.org/cgi-bin/translate/dna_aa"
- Attempt 4: curl -d "pre_text=cgtatgctaataccatgttccgcgtataacccagccgccagttccgctggcggcatttta&Submit=TRANSLATE SEQUENCE" http://web.expasy.org/cgi-bin/translate/dna_aa
Study the curl'ed Code
Below, going line by line clicking each link and describing where it leads
- http://www.w3.org/TR/html4/loose.dtd : Described as "the HTML 4.01 Transitional DTD, which includes presentation attributes and elements that W3C expects to phase out as support for style sheets matures."
- http://web.expasy.org/favicon.ico : The logo for Expasy.
- http://www.sib.swiss : The main web page for Swiss Institute of Bioinformatics.
- https://www.expasy.org : The Expasy Bioinformatics Resource Portal.
- http://web.expasy.org/translate/ : Expasy translation tool.
- https://en.wikipedia.org/wiki/Open_reading_frame : Wikipedia link to what an "open reading frame" is described as.
- https://www.expasy.org/disclaimer.html : Expasy disclaimer.
Identifiers:
- sib_top: The top of the webpage.
- sib_container: The "container" of the webpage.
- sib_header_small: The small header for the webpage.
- sib_body: The "body" of the webpage outside of the container.
- sib_footer: The footer at the bottom of the webpage.
- sib_expasy_logo: Logo at the top right of the page.
- resource_header: Header of the webpage.
- sib_header_nav: Homepage link at the top right of the page for navigation.
- Couldn't figure out how to use the grep and sed commands in command line.