Difference between revisions of "Emmatyrnauer Week 6"
From LMU BioDB 2017
Emmatyrnauer (talk | contribs) (answering more questions) |
Emmatyrnauer (talk | contribs) (adding template of links) |
||
| (10 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | #( | + | ==Questions== |
| − | + | #(See figure) [[File:Question_1_Assignment_6_Emma_Tyrnauer.png|400px|thumb|right|Question 1 Answer]] | |
| − | + | #The ratio is calculated by dividing one gene’s activity in cells gradually consuming all the available oxygen by its activity in control cells with unlimited oxygen. Therefore, Gene X goes from black to dim red to black to medium green. Gene Y goes from black to medium red to dim green to bright green. Finally, Gene Z goes from black to dim red and stays at dim red (in the order of 1 hour, 2 hour, 3 hour, and 4 hour, respectively). | |
| − | # | + | #Genes X and Y were transcribed similarly (had similar ratios). |
| − | + | #At time 0, no/very little change has occurred in terms of expression or repression of the gene because the cells have just been exposed to the experimental growth conditions. Spots with equal expression and repression compared to the control sample appear yellow because red and green mix to make yellow. | |
| − | # | + | #TEF4 was repressed over the course of the experiment indicated by the green color. TEF4's change in expression was part of the cell's response to a reduction in available glucose. Glucose is the source of energy for cells and in the absence of glucose, actions that expend the cells energy (such as translation) may be limited. |
| − | + | #The TCA cycle does not use glucose directly--glucose is converted into pyruvic acid. When glucose levels are low, pyruvic acid is modified and converted into acetyl-CoA, which then enters the TCA cycle. | |
| − | # | + | #These genes in the common pathway could all be induced or repressed by the same transcription factor (pg. 119). |
| − | # | + | #I would expect these spots to be red because since the repressor is absent, the cells would continue to express the gene even in the absence of glucose. |
| − | + | #I would expect the spots to be red since Yap1p target genes allow for increased resistance to environmental stresses. Therefore, a depletion of glucose in the environment (i.e. an environmental stress) would induce more expression of these genes. | |
| − | # | + | #Deletion of TUP1 and YAP1 would result in a red spot on the microarray for Tup1p compared to a green spot (expression compared to repression) and a brighter red spot for Yap1p (indicating more expression). |
| − | + | ||
| − | # | + | {{Emmatyrnauer}} |
| − | + | ||
| − | # | + | ==Acknowledgments== |
| − | + | #I worked with my homework partner [[User:ArashLari|Arash Lari]] over text. We were available for one another for questions about the assignment. | |
| − | + | #While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source. | |
| − | # | + | [[User:Emmatyrnauer|Emmatyrnauer]] ([[User talk:Emmatyrnauer|talk]]) 21:27, 9 October 2017 (PDT) |
| − | # | + | |
| − | # | + | ==References== |
| + | #LMU BioDB 2017. (2017). Week 6. Retrieved October 9, 2017, from https://xmlpipedb.cs.lmu.edu/biodb/fall2017/index.php/Week_6 | ||
| + | #Campbell, A.M. and Heyer, L.J. (2003), “Chapter 4: Basic Research with DNA Microarrays”, in Discovering Genomics, Proteomics, and Bioinformatics, Cold Spring Harbor Laboratory Press, pp. 107-124. | ||
| + | |||
| + | [[Category:Journal Entry]] | ||
Latest revision as of 06:11, 10 October 2017
Questions
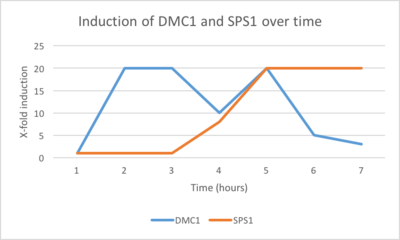
- (See figure)
- The ratio is calculated by dividing one gene’s activity in cells gradually consuming all the available oxygen by its activity in control cells with unlimited oxygen. Therefore, Gene X goes from black to dim red to black to medium green. Gene Y goes from black to medium red to dim green to bright green. Finally, Gene Z goes from black to dim red and stays at dim red (in the order of 1 hour, 2 hour, 3 hour, and 4 hour, respectively).
- Genes X and Y were transcribed similarly (had similar ratios).
- At time 0, no/very little change has occurred in terms of expression or repression of the gene because the cells have just been exposed to the experimental growth conditions. Spots with equal expression and repression compared to the control sample appear yellow because red and green mix to make yellow.
- TEF4 was repressed over the course of the experiment indicated by the green color. TEF4's change in expression was part of the cell's response to a reduction in available glucose. Glucose is the source of energy for cells and in the absence of glucose, actions that expend the cells energy (such as translation) may be limited.
- The TCA cycle does not use glucose directly--glucose is converted into pyruvic acid. When glucose levels are low, pyruvic acid is modified and converted into acetyl-CoA, which then enters the TCA cycle.
- These genes in the common pathway could all be induced or repressed by the same transcription factor (pg. 119).
- I would expect these spots to be red because since the repressor is absent, the cells would continue to express the gene even in the absence of glucose.
- I would expect the spots to be red since Yap1p target genes allow for increased resistance to environmental stresses. Therefore, a depletion of glucose in the environment (i.e. an environmental stress) would induce more expression of these genes.
- Deletion of TUP1 and YAP1 would result in a red spot on the microarray for Tup1p compared to a green spot (expression compared to repression) and a brighter red spot for Yap1p (indicating more expression).
Links
- My User Page
- List of Assignments
- List of Journal Entries
- List of Shared Journal Entries
- Class Journal Week 1
- Class Journal Week 2
- Class Journal Week 3
- Class Journal Week 4
- Class Journal Week 5
- Class Journal Week 6
- Class Journal Week 7
- Class Journal Week 8
- Class Journal Week 9
- Class Journal Week 10
- Group Journal Week 11
- Group Journal Week 12
- no week 13
- Group Journal Week 14 (executive summary)
- Group Journal Week 14 (executive summary)
- Group Journal Week 15 (executive summary)
Acknowledgments
- I worked with my homework partner Arash Lari over text. We were available for one another for questions about the assignment.
- While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source.
Emmatyrnauer (talk) 21:27, 9 October 2017 (PDT)
References
- LMU BioDB 2017. (2017). Week 6. Retrieved October 9, 2017, from https://xmlpipedb.cs.lmu.edu/biodb/fall2017/index.php/Week_6
- Campbell, A.M. and Heyer, L.J. (2003), “Chapter 4: Basic Research with DNA Microarrays”, in Discovering Genomics, Proteomics, and Bioinformatics, Cold Spring Harbor Laboratory Press, pp. 107-124.