Difference between revisions of "ArashLari Week 9"
(Pre-test information) |
(→Electronic Workbook: clarifying what passed means) |
||
| (10 intermediate revisions by the same user not shown) | |||
| Line 5: | Line 5: | ||
*21-genes_31-edges_Schade-data_estimation_output.graphml | *21-genes_31-edges_Schade-data_estimation_output.graphml | ||
Then proceed to the GRNSight-beta from the link provided in the instructions for this assignment | Then proceed to the GRNSight-beta from the link provided in the instructions for this assignment | ||
| + | *Note: A test passing means that it satisfied all the criteria detailed in the instructions | ||
== Test 1 == | == Test 1 == | ||
| + | Instructions: | ||
| + | *Load Graph - File Menu -> Open | ||
| + | *Restrict Graph to Viewport - Check | ||
| + | *Hide/Show Edge Weights - Select "Show With Mouse Over" | ||
| + | Result: '''PASSED''' | ||
== Test 2 == | == Test 2 == | ||
| + | Instructions: | ||
| + | *Load Graph - File Menu -> Import SIF | ||
| + | *Restrict Graph to Viewport - Check | ||
| + | *Hide/Show Edge Weights - Select "Show With Mouse Over" | ||
| + | Result: '''PASSED''' | ||
== Test 3 == | == Test 3 == | ||
| + | Instructions: | ||
| + | *Load Graph - File Menu -> Import GraphML | ||
| + | *Restrict Graph to Viewport - Check | ||
| + | *Hide/Show Edge Weights - Select "Show With Mouse Over" | ||
| + | Result: '''PASSED''' | ||
== Test 4 == | == Test 4 == | ||
| + | Instructions: | ||
| + | *Load Graph - File Menu -> Open | ||
| + | *Restrict Graph to Viewport - Uncheck | ||
| + | *Hide/Show Edge Weights - Select "Show With Mouse Over" | ||
| + | Result: '''PASSED''' | ||
== Test 5 == | == Test 5 == | ||
| + | Instructions: | ||
| + | *Load Graph - File Menu -> Import SIF | ||
| + | *Restrict Graph to Viewport - Uncheck | ||
| + | *Hide/Show Edge Weights - Select "Show With Mouse Over" | ||
| + | Result: '''PASSED''' | ||
== Test 6 == | == Test 6 == | ||
| + | Instructions: | ||
| + | *Load Graph - File Menu -> Import GraphML | ||
| + | *Restrict Graph to Viewport - Uncheck | ||
| + | *Hide/Show Edge Weights - Select "Show With Mouse Over" | ||
| + | Result: '''PASSED''' | ||
== Test 7 == | == Test 7 == | ||
| + | Instructions: | ||
| + | *Load Graph - File Menu -> Open | ||
| + | *Restrict Graph to Viewport - Check | ||
| + | *Hide/Show Edge Weights - Select "Always Show Edge Weights" | ||
| + | Result: '''PASSED''' | ||
== Test 8 == | == Test 8 == | ||
| + | Instructions: | ||
| + | *Load Graph - File Menu -> Import SIF | ||
| + | *Restrict Graph to Viewport - Check | ||
| + | *Hide/Show Edge Weights - Select "Always Show Edge Weights" | ||
| + | Result: '''PASSED''' | ||
== Test 9 == | == Test 9 == | ||
| + | Instructions: | ||
| + | *Load Graph - File Menu -> Import GraphML | ||
| + | *Restrict Graph to Viewport - Check | ||
| + | *Hide/Show Edge Weights - Select "Always Show Edge Weights" | ||
| + | Result: '''PASSED''' | ||
== Test 10 == | == Test 10 == | ||
| + | Instructions: | ||
| + | *Load Graph - File Menu -> Open | ||
| + | *Restrict Graph to Viewport - Uncheck | ||
| + | *Hide/Show Edge Weights - Select "Always Show Edge Weights" | ||
| + | Result: '''PASSED''' | ||
== Test 11 == | == Test 11 == | ||
| + | Instructions: | ||
| + | *Load Graph - File Menu -> Import SIF | ||
| + | *Restrict Graph to Viewport - Uncheck | ||
| + | *Hide/Show Edge Weights - Select "Always Show Edge Weights" | ||
| + | Result: '''PASSED''' | ||
== Test 12 == | == Test 12 == | ||
| + | Instructions: | ||
| + | *Load Graph - File Menu -> Import GraphML | ||
| + | *Restrict Graph to Viewport - Uncheck | ||
| + | *Hide/Show Edge Weights - Select "Always Show Edge Weights" | ||
| + | Result: '''PASSED''' | ||
== Test 13 == | == Test 13 == | ||
| + | Instructions: | ||
| + | *Load Graph - File Menu -> Open | ||
| + | *Restrict Graph to Viewport - Check | ||
| + | *Hide/Show Edge Weights - Select "Never Show Edge Weights" | ||
| + | Result: '''PASSED''' | ||
== Test 14 == | == Test 14 == | ||
| + | Instructions: | ||
| + | *Load Graph - File Menu -> Import SIF | ||
| + | *Restrict Graph to Viewport - Check | ||
| + | *Hide/Show Edge Weights - Select "Never Show Edge Weights" | ||
| + | Result: '''PASSED''' | ||
== Test 15 == | == Test 15 == | ||
| + | Instructions: | ||
| + | *Load Graph - File Menu -> Import GraphML | ||
| + | *Restrict Graph to Viewport - Check | ||
| + | *Hide/Show Edge Weights - Select "Never Show Edge Weights" | ||
| + | Result: '''PASSED''' | ||
== Test 16 == | == Test 16 == | ||
| + | Instructions: | ||
| + | *Load Graph - File Menu -> Open | ||
| + | *Restrict Graph to Viewport - Uncheck | ||
| + | *Hide/Show Edge Weights - Select "Never Show Edge Weights" | ||
| + | Result: '''PASSED''' | ||
== Test 17 == | == Test 17 == | ||
| + | Instructions: | ||
| + | *Load Graph - File Menu -> Import SIF | ||
| + | *Restrict Graph to Viewport - Uncheck | ||
| + | *Hide/Show Edge Weights - Select "Never Show Edge Weights" | ||
| + | Result: '''PASSED''' | ||
== Test 18 == | == Test 18 == | ||
| + | Instructions: | ||
| + | *Load Graph - File Menu -> Import GraphML | ||
| + | *Restrict Graph to Viewport - Uncheck | ||
| + | *Hide/Show Edge Weights - Select "Never Show Edge Weights" | ||
| + | Result: '''PASSED''' | ||
| + | == Observations == | ||
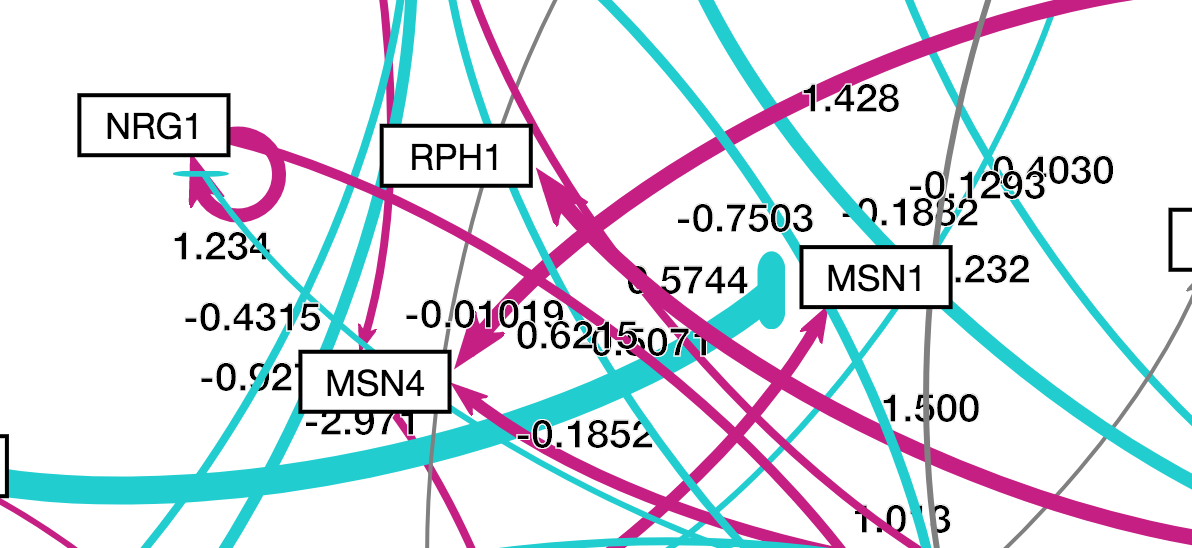
| + | While they all passed, it should be noted that for tests 7 through 12, the user can technically see all the numbers, but sometimes they might overlap and become hard to read. | ||
| + | This can be seen in the following image: | ||
| − | = | + | [[File:ArashObservation.png]] |
| − | curl 'http://rest.ensembl.org/lookup/symbol/saccharomyces_cerevisiae/ | + | |
| + | = Web Service API Exploration = | ||
| + | While I was originally confused as to what the assignment was asking for, once we figured it out it became much clearer. I simply went to the ensemble api page, and found the command that returned the json file using the gene name. The example given on the website had the species as "homo_sapien", so i changed that to "saccharomyces_cerevisiae" in the url and put the name of a random yeast file to test it. This process worked, but because the instructions pointed to [http://rest.ensembl.org/documentation/info/lookup] I used the ID retrieved from the first command and placed it in the command given from this page to do it the way the instructions alluded to. | ||
| + | ==Deliverable== | ||
| + | *Note: to find the json file for any other gene, simply replace the text that says "GENE" in the url with your desired gene name. (this code won't work as it's generic) | ||
| + | curl 'http://rest.ensembl.org/lookup/symbol/saccharomyces_cerevisiae/GENE?' -H 'Content-type:application/json' | ||
| + | From there you could put the id retrieved from the json file into the url where it says "ID" in uppercase, but the json file above also returns all the relevant information already. | ||
| + | curl 'http://rest.ensembl.org/lookup/id/ID?expand=1' -H 'Content-type:application/json' | ||
= Acknowledgements = | = Acknowledgements = | ||
| − | + | Nicole Kalcic was my partner and we met up once and worked over text message. I also worked with Zach Van Ysseldyk and Antoni Porras on the Web API section and we figured out how to return the desired deliverable together. | |
| + | While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source. | ||
[[User:ArashLari|ArashLari]] ([[User talk:ArashLari|talk]]) 15:36, 30 October 2017 (PDT) | [[User:ArashLari|ArashLari]] ([[User talk:ArashLari|talk]]) 15:36, 30 October 2017 (PDT) | ||
{{Template:ArashLari}} | {{Template:ArashLari}} | ||
Latest revision as of 06:12, 31 October 2017
Electronic Workbook
For the purposes of testing I used the following files:
- 21-genes_31-edges_Schade-data_estimation_output.xlsx
- 21-genes_31-edges_Schade-data_estimation_output.sif
- 21-genes_31-edges_Schade-data_estimation_output.graphml
Then proceed to the GRNSight-beta from the link provided in the instructions for this assignment
- Note: A test passing means that it satisfied all the criteria detailed in the instructions
Test 1
Instructions:
- Load Graph - File Menu -> Open
- Restrict Graph to Viewport - Check
- Hide/Show Edge Weights - Select "Show With Mouse Over"
Result: PASSED
Test 2
Instructions:
- Load Graph - File Menu -> Import SIF
- Restrict Graph to Viewport - Check
- Hide/Show Edge Weights - Select "Show With Mouse Over"
Result: PASSED
Test 3
Instructions:
- Load Graph - File Menu -> Import GraphML
- Restrict Graph to Viewport - Check
- Hide/Show Edge Weights - Select "Show With Mouse Over"
Result: PASSED
Test 4
Instructions:
- Load Graph - File Menu -> Open
- Restrict Graph to Viewport - Uncheck
- Hide/Show Edge Weights - Select "Show With Mouse Over"
Result: PASSED
Test 5
Instructions:
- Load Graph - File Menu -> Import SIF
- Restrict Graph to Viewport - Uncheck
- Hide/Show Edge Weights - Select "Show With Mouse Over"
Result: PASSED
Test 6
Instructions:
- Load Graph - File Menu -> Import GraphML
- Restrict Graph to Viewport - Uncheck
- Hide/Show Edge Weights - Select "Show With Mouse Over"
Result: PASSED
Test 7
Instructions:
- Load Graph - File Menu -> Open
- Restrict Graph to Viewport - Check
- Hide/Show Edge Weights - Select "Always Show Edge Weights"
Result: PASSED
Test 8
Instructions:
- Load Graph - File Menu -> Import SIF
- Restrict Graph to Viewport - Check
- Hide/Show Edge Weights - Select "Always Show Edge Weights"
Result: PASSED
Test 9
Instructions:
- Load Graph - File Menu -> Import GraphML
- Restrict Graph to Viewport - Check
- Hide/Show Edge Weights - Select "Always Show Edge Weights"
Result: PASSED
Test 10
Instructions:
- Load Graph - File Menu -> Open
- Restrict Graph to Viewport - Uncheck
- Hide/Show Edge Weights - Select "Always Show Edge Weights"
Result: PASSED
Test 11
Instructions:
- Load Graph - File Menu -> Import SIF
- Restrict Graph to Viewport - Uncheck
- Hide/Show Edge Weights - Select "Always Show Edge Weights"
Result: PASSED
Test 12
Instructions:
- Load Graph - File Menu -> Import GraphML
- Restrict Graph to Viewport - Uncheck
- Hide/Show Edge Weights - Select "Always Show Edge Weights"
Result: PASSED
Test 13
Instructions:
- Load Graph - File Menu -> Open
- Restrict Graph to Viewport - Check
- Hide/Show Edge Weights - Select "Never Show Edge Weights"
Result: PASSED
Test 14
Instructions:
- Load Graph - File Menu -> Import SIF
- Restrict Graph to Viewport - Check
- Hide/Show Edge Weights - Select "Never Show Edge Weights"
Result: PASSED
Test 15
Instructions:
- Load Graph - File Menu -> Import GraphML
- Restrict Graph to Viewport - Check
- Hide/Show Edge Weights - Select "Never Show Edge Weights"
Result: PASSED
Test 16
Instructions:
- Load Graph - File Menu -> Open
- Restrict Graph to Viewport - Uncheck
- Hide/Show Edge Weights - Select "Never Show Edge Weights"
Result: PASSED
Test 17
Instructions:
- Load Graph - File Menu -> Import SIF
- Restrict Graph to Viewport - Uncheck
- Hide/Show Edge Weights - Select "Never Show Edge Weights"
Result: PASSED
Test 18
Instructions:
- Load Graph - File Menu -> Import GraphML
- Restrict Graph to Viewport - Uncheck
- Hide/Show Edge Weights - Select "Never Show Edge Weights"
Result: PASSED
Observations
While they all passed, it should be noted that for tests 7 through 12, the user can technically see all the numbers, but sometimes they might overlap and become hard to read. This can be seen in the following image:
Web Service API Exploration
While I was originally confused as to what the assignment was asking for, once we figured it out it became much clearer. I simply went to the ensemble api page, and found the command that returned the json file using the gene name. The example given on the website had the species as "homo_sapien", so i changed that to "saccharomyces_cerevisiae" in the url and put the name of a random yeast file to test it. This process worked, but because the instructions pointed to [1] I used the ID retrieved from the first command and placed it in the command given from this page to do it the way the instructions alluded to.
Deliverable
- Note: to find the json file for any other gene, simply replace the text that says "GENE" in the url with your desired gene name. (this code won't work as it's generic)
curl 'http://rest.ensembl.org/lookup/symbol/saccharomyces_cerevisiae/GENE?' -H 'Content-type:application/json'
From there you could put the id retrieved from the json file into the url where it says "ID" in uppercase, but the json file above also returns all the relevant information already.
curl 'http://rest.ensembl.org/lookup/id/ID?expand=1' -H 'Content-type:application/json'
Acknowledgements
Nicole Kalcic was my partner and we met up once and worked over text message. I also worked with Zach Van Ysseldyk and Antoni Porras on the Web API section and we figured out how to return the desired deliverable together. While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source. ArashLari (talk) 15:36, 30 October 2017 (PDT) Arash Lari
BIOL/CMSI 367-01: Biological Databases Fall 2017
Assignments
- Week 1
- Week 2
- Week 3
- Week 4
- Week 5
- Week 6
- Week 7
- Week 8
- Week 9
- Week 10
- Week 11
- Week 12
- Week 13
- Week 14
- Week 15
- Week 16
Journal Entries:
- ArashLari Week 2
- ArashLari Week 3
- ArashLari Week 4
- ArashLari Week 5
- ArashLari Week 6
- ArashLari Week 7
- ArashLari Week 8
- ArashLari Week 9
- ArashLari Week 10
- ArashLari Week 11
- ArashLari Week 12
- ArashLari Week 13
- ArashLari Week 14
- ArashLari Week 15
- ArashLari Week 16
Shared Journals:
- ArashLari Week 1 Journal
- ArashLari Week 2 Journal
- ArashLari Week 3 Journal
- ArashLari Week 4 Journal
- ArashLari Week 5 Journal
- ArashLari Week 6 Journal
- ArashLari Week 7 Journal
- ArashLari Week 8 Journal
- ArashLari Week 9 Journal
- ArashLari Week 10 Journal
- ArashLari Week 11 Journal
- ArashLari Week 12 Journal
- ArashLari Week 13 Journal
- ArashLari Week 14 Journal
- ArashLari Week 15 Journal
- ArashLari Week 16 Journal
- Page Desiigner