Difference between revisions of "Cwong34 Week 12"
From LMU BioDB 2017
(Added link to article) |
(Changed name of presentation link) |
||
| (10 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
=Article= | =Article= | ||
Sahara, T., Goda, T., & Ohgiya, S. (2002). [http://www.jbc.org/content/277/51/50015.long Comprehensive expression analysis of time-dependent genetic responses in yeast cells to low temperature]. Journal of Biological Chemistry, 277(51), 50015-50021. | Sahara, T., Goda, T., & Ohgiya, S. (2002). [http://www.jbc.org/content/277/51/50015.long Comprehensive expression analysis of time-dependent genetic responses in yeast cells to low temperature]. Journal of Biological Chemistry, 277(51), 50015-50021. | ||
| + | =Presentation= | ||
| + | [[Media:Cold_Shock_Yeast_Genome_Response.pdf|Journal Club Presentation 2]] | ||
| + | |||
| + | =Flow Chart= | ||
| + | [[Image:Sahara_T_et_al_article_flowchart_CW.png]] | ||
=Outline= | =Outline= | ||
| + | ==Experiment design and procedures== | ||
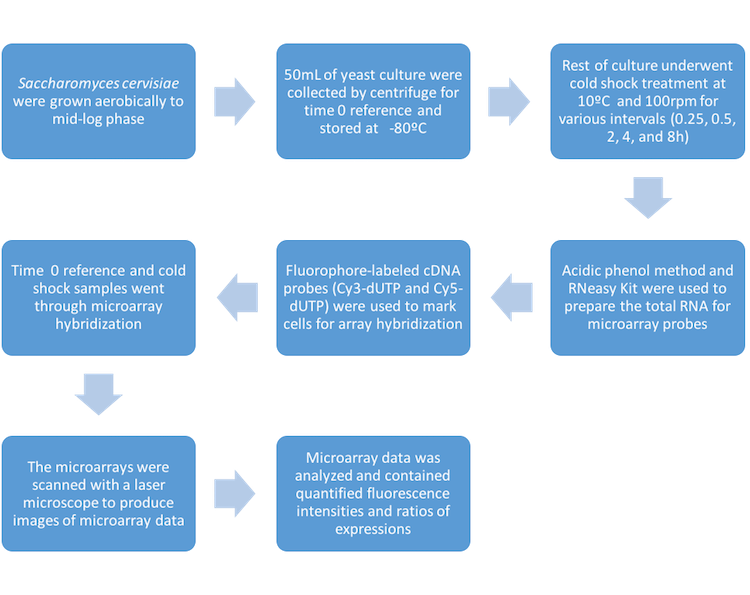
| + | *Purchased cDNA microarray of ''Saccharomyces cervisiae'' from DNA Chip Research Inc. | ||
| + | *Used ''S. cervisiae''YPH500 | ||
| + | *Grew yeast cells aerobically in YPD medium at 30 degrees celsius and shaken at 100rpm. | ||
| + | *YPD is made up of: | ||
| + | **1% yeast extract | ||
| + | **2% peptone | ||
| + | **2% glucose | ||
| + | *Yeast cells were grown to "mid-log phase" where they were still maturing, but not fully reproducing | ||
| + | *50mL of culture was taken and centrifuged to collect the cells and be used as a time 0 reference for the rest of the experiment | ||
| + | *The time 0 reference cells were stored at -80 degrees celsius | ||
| + | *The rest of culture was used for the experimental samples and was cold shocked at 10 degrees celsius | ||
| + | *Samples were collected at various times: 0.25, 0.5, 2, 4, and 8h | ||
| + | *Acidic phenol method and RNeasy Mini Kit were used to prepare the RNA | ||
| + | *The RNA was used to prepare fluorophore-labeled cDNA probes | ||
| + | *These probes marked the cells for array hybridization | ||
| + | *The microarrays were scanned with a laser microscope and were analyzed | ||
| + | *Repeated process twice | ||
| + | *Averaged the expression ratios of the separate experiments for final data | ||
| + | ==Results & discussion== | ||
| + | *Analyzed the microarray of cDNAs of 5,803 genes in yeast genome | ||
| + | *There was a diauxic shift in cells that experienced cold shock | ||
| + | **Down-shift of some diauxic shift-inducible genes in late phase of cold shock | ||
| + | *Low temperature affects expression of ~25% of the yeast genes | ||
| + | *Number of up-regulated genes increased from 41 at 0.25h to 536 at 8h | ||
| + | *Number of down-regulated genes also increased from 4 at 0.25h to 488 at 8h | ||
| + | *Gene expression changes significantly in both ways (up & down-regulation) to adapt cells to colder environment, similar to reactions to other stresses, like heat, salinity, hydrogen peroxide, and osmotic stresses | ||
| + | *Table 1 shows the number of genes that significantly changed expression (2-fold or more), up-regulating or down-regulating | ||
| + | ===Clustering analysis=== | ||
| + | *Clustering of genes (genes that were close together) were analyzed in the ways they responded | ||
| + | *Genes separated into 5 different clusters (Fig. 1): | ||
| + | **1A: Unclassified proteins | ||
| + | **1B: Amino acid biosynthesis and metabolism | ||
| + | **1C: RNA Polymerase I & RNA processing | ||
| + | **1D: Ribosomal proteins | ||
| + | **1E: Not labeled | ||
| + | *Shows cooperative regulation | ||
| + | **1C: up-regulated in early phase (0-0.5h), then down-regulated in late phase (4-8h) | ||
| + | **1D & 1E: up-regulated in mid-late phase (2h & 4-8h) | ||
| + | ===Transcription related genes=== | ||
| + | *Looked at clusters of genes relating to RNA polymerase I & RNA processing | ||
| + | **All up-regulated in early phase | ||
| + | *Cooperative regulation of genes involved in transcription (Fig. 2) | ||
| + | *2 clusters of these genes: | ||
| + | **Down-regulating (2A, 2B, & 2D) | ||
| + | ***2A (RNA polymerase I & RNA processing) & 2B (rRNA processing) up-regulated in early phase, then down regulated in late phase | ||
| + | ***2D: mRNA transcription | ||
| + | **Up-regulating (2C) | ||
| + | ***2C: mRNA transcription | ||
| + | ***High up-regulating in mid phase | ||
| + | *Up-regulated genes that had to do with basic transcriptional functions, like genes encoding for regulatory proteins for amino acid production | ||
| + | *Down-regulated genes were not essential for basic life, like genes encoding heat shock transcription factor or a transcription factor for drug resistant genes | ||
| + | *Factors essential to transcription & processing of rRNAs were up-regulated | ||
| + | *Genes for synthesis and transcription regulation of mRNAs had mix responses | ||
| + | ===Ribosomal protein related genes=== | ||
| + | *Genes separated into four clusters (Fig. 3): | ||
| + | **3A & 3B: cytosolic ribosome | ||
| + | **3C: translational control factors | ||
| + | **3D: tRNA syntheatases | ||
| + | *3A up-regulated in early-mid phase, then down-regulated in late phase | ||
| + | *3B up-regulated in early-mid phase, and slightly down-regulated in the late phase | ||
| + | *3C continuously up-regulated starting in the early phase | ||
| + | *3D overall down-regulated | ||
| + | *Genes encoding ribosomal proteins = most of up-regulated genes at 2h | ||
| + | *Almost all up-regulating ribosomal proteins had similar expressions, showing cooperative regulation | ||
| + | *Low temperature impairs translational ability | ||
| + | *When genes related to ribosomal proteins and rRNA processing don't work, there is cold sensitivity | ||
| + | *Yeast up-regulates genes encoding ribosomal proteins, so compensates for less efficient/productive translation to overcome cold sensitivity | ||
| + | ===Cell rescue, defense, death, and aging genes=== | ||
| + | *Genes separated into four clusters (Fig.. 4): | ||
| + | **4A: not labeled | ||
| + | **4B & 4C: stress response - high up-regulation in mid-late phase | ||
| + | **4D: stress response and chaperone - high down-regulation in mid-late phase | ||
| + | *Heat shock protein genes (HSPs) were down-regulated in low temperatures, except HSP12 and HSP26 | ||
| + | *This may mean HSP12 and HSP26 have different transcription regulation than other HSPs | ||
| + | *Graumann, et al., 1996 did similar experiment with Bacillus subtilis and found peptidyl prolyl cis/trans isomerases were induced in low temperatures, so protein folding genes were up-regulated | ||
| + | ===Metabolism and energy production genes=== | ||
| + | *8 clusters (Fig. 5): | ||
| + | **5A: nucleotide metabolism - up-regulated early-mid phase, then down-regulated | ||
| + | **5B & 5E: not annotated | ||
| + | **5C & 5D: C-compound and carbohydrate metabolism - up-regulated | ||
| + | **5F & 5H: amino acid metabolism - down-regulated | ||
| + | **5G: C-compound and carbohydrate utilization - down-regulated | ||
| + | *Clusters show a lot of cooperative regulation between glycogen and trehalose biosynthesis genes | ||
| + | **glycogen and trehalose both reserve carbohydrates | ||
| + | *Genes involved in glycogen production up-regulated in mid-late phases | ||
| + | *Curious that glycogen degradation gene (GPH1) was also up-regulated | ||
| + | *Tests in heat shock found even when glycogen levels were low, yeast still recylced/degraded it | ||
| + | *Trehalose may help protect cellular membrane, which may help to keep yeast cells intact at low temperatures | ||
| + | ===Signal transduction genes=== | ||
| + | *Clusters (Fig. 6): | ||
| + | **6A - down-regulated | ||
| + | **6B - little up-regulation in early phase, then down-regulation | ||
| + | **6C - little down-regulation in early phase, then up-regulation in late phase | ||
| + | *Clusters were not defined in the article | ||
| + | *20% of signal transduction genes changed expression significantly | ||
| + | *Particularly genes related to cAMP-PKA pathway were up-regulated | ||
| + | **increased signaling | ||
| + | *Increase in PKA signaling known to be response to stresses and controlled by Msn2p/4p transcription factors | ||
| + | *cAMP-PKA pathway involved in: | ||
| + | **metabolism control | ||
| + | **stress resistance | ||
| + | **cell proliferation | ||
| + | *Many Msn2p/4p genes up-regulated (50% of them), which had roles in: | ||
| + | **glycogen synthesis | ||
| + | **trehalose synthesis | ||
| + | **stress resistance | ||
| + | *Heat shock gene promoters are suppressed by Msn2p/4p | ||
| + | *Cooperative regulation in genes of Msn2p/4p and cAMP-PKA pathway that are related to stress response and biosynthesis of glycogen and trehalose | ||
| + | *cAMP-PKA pathway may have effect on yeast gene expression after cold shock | ||
| + | ==Conclusion== | ||
| + | *Clustering analysis shows different responses in three phases: | ||
| + | **Early phase (0-2h) - transcription (RNA polymerase I & rRNA processing) related genes up-regulated | ||
| + | **Middle phase (2-4h) - ribosomal protein related genes up-regulated | ||
| + | **Late phase (4-8h) - stress response genes up-regulated | ||
| + | *This shows adaptation to environment (low temperature) involves three sequential events (the three phases) | ||
| + | *RNA polymerase I & rRNA processing genes respond first by up-regulating because help transcription and translation, which become less efficient when exposed to low temperature | ||
| + | *Ribosomal protein genes up-regulated in middle phase to further assist in maintaining translational ability | ||
| + | *Maintaining translation is priority to yeast in cold shock because they need to make proteins to help maintain integrity and basic functions of cells | ||
| + | *Stress response induced genes follow in up-regulation, so next step in yeast cells is to adapt and gain tolerance to low temperature | ||
| + | =Terms= | ||
| + | #Nucleolin: a protein associated with nucleolar in growing eukaryotic cells (NCBI, 2017). | ||
| + | #Hypoxia: low levels of oxygen in body tissues (Hine & Martin, 2015). | ||
| + | #Ubiquitin: a protein that marks which proteins are going to be broken down (Hine & Martin, 2015). | ||
| + | #"Mid-log" or "lag" phase: bacterial cells are maturing, but they have not yet reached their maximum reproduction rate (Hine & Martin, 2015). | ||
| + | #Diauxic shift: the switch of a microorganism from using one type of sugar to using another (King, et al., 2014). | ||
| + | #Cytosolic: contents of the fluid in the cytoplasm of a cell (King, et al., 2014). | ||
| + | #Peptidyl prolyl cis/trans isomerases: Enzymes that changes conformation of prolyl bonds to cis or trans, chaning the tertiary structure of a protein (Lackie, 2013). | ||
| + | #cAMP-PKA pathway: the activation of PKA by cAMP (Lackie, 2013). | ||
| + | #GTPase: enzymes that hydrolyze GTP (Lackie, 2013). | ||
| + | #Phosphatase: an enzyme that helps remove a phosphate group from an organic compound (Hine & Martin, 2015). | ||
=Acknowledgments= | =Acknowledgments= | ||
| + | #I met with Dina outside of class, and we worked on our presentation together. | ||
| + | |||
| + | While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source. [[User:Cwong34|Cwong34]] ([[User talk:Cwong34|talk]]) 17:28, 20 November 2017 (PST) | ||
| + | |||
=References= | =References= | ||
| + | #Graumann, P., Schröder, K., Schmid, R., & Marahiel, M.A. (1996). Cold shock stress-induced proteins in Bacillus subtilis. ''Journal of Bacteriology'', 178(15), 4611-4619. doi: 10.1128/jb.178.15.4611-4619.1996 | ||
| + | #Hine, R. & Martin, E. (Eds.). (2015). ''A Dictionary of Biology''. In ''Oxford Reference''. Retrieved from http://www.oxfordreference.com/view/10.1093/acref/9780198714378.001.0001/acref-9780198714378 | ||
| + | #King, R.C., Mulligan, P.K., & Stansfield, W.D. (Eds.). (2014). ''A Dictionary of Genetics''. In ''Oxford Reference''. Retrieved from http://www.oxfordreference.com/view/10.1093/acref/9780199766444.001.0001/acref-9780199766444 | ||
| + | #Lackie, J.L. (2013). ''The Dictionary of Cell and Molecular Biology''. In ''Science Direct''. Retrieved from http://www.sciencedirect.com/science/book/9780123849311 | ||
#LMU BioDB 2017. (2017). Week 12. Retrieved November 14, 2017, from https://xmlpipedb.cs.lmu.edu/biodb/fall2017/index.php/Week_12 | #LMU BioDB 2017. (2017). Week 12. Retrieved November 14, 2017, from https://xmlpipedb.cs.lmu.edu/biodb/fall2017/index.php/Week_12 | ||
| + | #NCBI. (2017). Nucleolin. Retrieved November 20, 2017, from http://www.uniprot.org/uniprot/P19338 | ||
#Sahara, T., Goda, T., & Ohgiya, S. (2002). Comprehensive expression analysis of time-dependent genetic responses in yeast cells to low temperature. Journal of Biological Chemistry, 277(51), 50015-50021. | #Sahara, T., Goda, T., & Ohgiya, S. (2002). Comprehensive expression analysis of time-dependent genetic responses in yeast cells to low temperature. Journal of Biological Chemistry, 277(51), 50015-50021. | ||
Latest revision as of 08:05, 21 November 2017
Contents
Article
Sahara, T., Goda, T., & Ohgiya, S. (2002). Comprehensive expression analysis of time-dependent genetic responses in yeast cells to low temperature. Journal of Biological Chemistry, 277(51), 50015-50021.
Presentation
Flow Chart
Outline
Experiment design and procedures
- Purchased cDNA microarray of Saccharomyces cervisiae from DNA Chip Research Inc.
- Used S. cervisiaeYPH500
- Grew yeast cells aerobically in YPD medium at 30 degrees celsius and shaken at 100rpm.
- YPD is made up of:
- 1% yeast extract
- 2% peptone
- 2% glucose
- Yeast cells were grown to "mid-log phase" where they were still maturing, but not fully reproducing
- 50mL of culture was taken and centrifuged to collect the cells and be used as a time 0 reference for the rest of the experiment
- The time 0 reference cells were stored at -80 degrees celsius
- The rest of culture was used for the experimental samples and was cold shocked at 10 degrees celsius
- Samples were collected at various times: 0.25, 0.5, 2, 4, and 8h
- Acidic phenol method and RNeasy Mini Kit were used to prepare the RNA
- The RNA was used to prepare fluorophore-labeled cDNA probes
- These probes marked the cells for array hybridization
- The microarrays were scanned with a laser microscope and were analyzed
- Repeated process twice
- Averaged the expression ratios of the separate experiments for final data
Results & discussion
- Analyzed the microarray of cDNAs of 5,803 genes in yeast genome
- There was a diauxic shift in cells that experienced cold shock
- Down-shift of some diauxic shift-inducible genes in late phase of cold shock
- Low temperature affects expression of ~25% of the yeast genes
- Number of up-regulated genes increased from 41 at 0.25h to 536 at 8h
- Number of down-regulated genes also increased from 4 at 0.25h to 488 at 8h
- Gene expression changes significantly in both ways (up & down-regulation) to adapt cells to colder environment, similar to reactions to other stresses, like heat, salinity, hydrogen peroxide, and osmotic stresses
- Table 1 shows the number of genes that significantly changed expression (2-fold or more), up-regulating or down-regulating
Clustering analysis
- Clustering of genes (genes that were close together) were analyzed in the ways they responded
- Genes separated into 5 different clusters (Fig. 1):
- 1A: Unclassified proteins
- 1B: Amino acid biosynthesis and metabolism
- 1C: RNA Polymerase I & RNA processing
- 1D: Ribosomal proteins
- 1E: Not labeled
- Shows cooperative regulation
- 1C: up-regulated in early phase (0-0.5h), then down-regulated in late phase (4-8h)
- 1D & 1E: up-regulated in mid-late phase (2h & 4-8h)
- Looked at clusters of genes relating to RNA polymerase I & RNA processing
- All up-regulated in early phase
- Cooperative regulation of genes involved in transcription (Fig. 2)
- 2 clusters of these genes:
- Down-regulating (2A, 2B, & 2D)
- 2A (RNA polymerase I & RNA processing) & 2B (rRNA processing) up-regulated in early phase, then down regulated in late phase
- 2D: mRNA transcription
- Up-regulating (2C)
- 2C: mRNA transcription
- High up-regulating in mid phase
- Down-regulating (2A, 2B, & 2D)
- Up-regulated genes that had to do with basic transcriptional functions, like genes encoding for regulatory proteins for amino acid production
- Down-regulated genes were not essential for basic life, like genes encoding heat shock transcription factor or a transcription factor for drug resistant genes
- Factors essential to transcription & processing of rRNAs were up-regulated
- Genes for synthesis and transcription regulation of mRNAs had mix responses
- Genes separated into four clusters (Fig. 3):
- 3A & 3B: cytosolic ribosome
- 3C: translational control factors
- 3D: tRNA syntheatases
- 3A up-regulated in early-mid phase, then down-regulated in late phase
- 3B up-regulated in early-mid phase, and slightly down-regulated in the late phase
- 3C continuously up-regulated starting in the early phase
- 3D overall down-regulated
- Genes encoding ribosomal proteins = most of up-regulated genes at 2h
- Almost all up-regulating ribosomal proteins had similar expressions, showing cooperative regulation
- Low temperature impairs translational ability
- When genes related to ribosomal proteins and rRNA processing don't work, there is cold sensitivity
- Yeast up-regulates genes encoding ribosomal proteins, so compensates for less efficient/productive translation to overcome cold sensitivity
Cell rescue, defense, death, and aging genes
- Genes separated into four clusters (Fig.. 4):
- 4A: not labeled
- 4B & 4C: stress response - high up-regulation in mid-late phase
- 4D: stress response and chaperone - high down-regulation in mid-late phase
- Heat shock protein genes (HSPs) were down-regulated in low temperatures, except HSP12 and HSP26
- This may mean HSP12 and HSP26 have different transcription regulation than other HSPs
- Graumann, et al., 1996 did similar experiment with Bacillus subtilis and found peptidyl prolyl cis/trans isomerases were induced in low temperatures, so protein folding genes were up-regulated
Metabolism and energy production genes
- 8 clusters (Fig. 5):
- 5A: nucleotide metabolism - up-regulated early-mid phase, then down-regulated
- 5B & 5E: not annotated
- 5C & 5D: C-compound and carbohydrate metabolism - up-regulated
- 5F & 5H: amino acid metabolism - down-regulated
- 5G: C-compound and carbohydrate utilization - down-regulated
- Clusters show a lot of cooperative regulation between glycogen and trehalose biosynthesis genes
- glycogen and trehalose both reserve carbohydrates
- Genes involved in glycogen production up-regulated in mid-late phases
- Curious that glycogen degradation gene (GPH1) was also up-regulated
- Tests in heat shock found even when glycogen levels were low, yeast still recylced/degraded it
- Trehalose may help protect cellular membrane, which may help to keep yeast cells intact at low temperatures
Signal transduction genes
- Clusters (Fig. 6):
- 6A - down-regulated
- 6B - little up-regulation in early phase, then down-regulation
- 6C - little down-regulation in early phase, then up-regulation in late phase
- Clusters were not defined in the article
- 20% of signal transduction genes changed expression significantly
- Particularly genes related to cAMP-PKA pathway were up-regulated
- increased signaling
- Increase in PKA signaling known to be response to stresses and controlled by Msn2p/4p transcription factors
- cAMP-PKA pathway involved in:
- metabolism control
- stress resistance
- cell proliferation
- Many Msn2p/4p genes up-regulated (50% of them), which had roles in:
- glycogen synthesis
- trehalose synthesis
- stress resistance
- Heat shock gene promoters are suppressed by Msn2p/4p
- Cooperative regulation in genes of Msn2p/4p and cAMP-PKA pathway that are related to stress response and biosynthesis of glycogen and trehalose
- cAMP-PKA pathway may have effect on yeast gene expression after cold shock
Conclusion
- Clustering analysis shows different responses in three phases:
- Early phase (0-2h) - transcription (RNA polymerase I & rRNA processing) related genes up-regulated
- Middle phase (2-4h) - ribosomal protein related genes up-regulated
- Late phase (4-8h) - stress response genes up-regulated
- This shows adaptation to environment (low temperature) involves three sequential events (the three phases)
- RNA polymerase I & rRNA processing genes respond first by up-regulating because help transcription and translation, which become less efficient when exposed to low temperature
- Ribosomal protein genes up-regulated in middle phase to further assist in maintaining translational ability
- Maintaining translation is priority to yeast in cold shock because they need to make proteins to help maintain integrity and basic functions of cells
- Stress response induced genes follow in up-regulation, so next step in yeast cells is to adapt and gain tolerance to low temperature
Terms
- Nucleolin: a protein associated with nucleolar in growing eukaryotic cells (NCBI, 2017).
- Hypoxia: low levels of oxygen in body tissues (Hine & Martin, 2015).
- Ubiquitin: a protein that marks which proteins are going to be broken down (Hine & Martin, 2015).
- "Mid-log" or "lag" phase: bacterial cells are maturing, but they have not yet reached their maximum reproduction rate (Hine & Martin, 2015).
- Diauxic shift: the switch of a microorganism from using one type of sugar to using another (King, et al., 2014).
- Cytosolic: contents of the fluid in the cytoplasm of a cell (King, et al., 2014).
- Peptidyl prolyl cis/trans isomerases: Enzymes that changes conformation of prolyl bonds to cis or trans, chaning the tertiary structure of a protein (Lackie, 2013).
- cAMP-PKA pathway: the activation of PKA by cAMP (Lackie, 2013).
- GTPase: enzymes that hydrolyze GTP (Lackie, 2013).
- Phosphatase: an enzyme that helps remove a phosphate group from an organic compound (Hine & Martin, 2015).
Acknowledgments
- I met with Dina outside of class, and we worked on our presentation together.
While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source. Cwong34 (talk) 17:28, 20 November 2017 (PST)
References
- Graumann, P., Schröder, K., Schmid, R., & Marahiel, M.A. (1996). Cold shock stress-induced proteins in Bacillus subtilis. Journal of Bacteriology, 178(15), 4611-4619. doi: 10.1128/jb.178.15.4611-4619.1996
- Hine, R. & Martin, E. (Eds.). (2015). A Dictionary of Biology. In Oxford Reference. Retrieved from http://www.oxfordreference.com/view/10.1093/acref/9780198714378.001.0001/acref-9780198714378
- King, R.C., Mulligan, P.K., & Stansfield, W.D. (Eds.). (2014). A Dictionary of Genetics. In Oxford Reference. Retrieved from http://www.oxfordreference.com/view/10.1093/acref/9780199766444.001.0001/acref-9780199766444
- Lackie, J.L. (2013). The Dictionary of Cell and Molecular Biology. In Science Direct. Retrieved from http://www.sciencedirect.com/science/book/9780123849311
- LMU BioDB 2017. (2017). Week 12. Retrieved November 14, 2017, from https://xmlpipedb.cs.lmu.edu/biodb/fall2017/index.php/Week_12
- NCBI. (2017). Nucleolin. Retrieved November 20, 2017, from http://www.uniprot.org/uniprot/P19338
- Sahara, T., Goda, T., & Ohgiya, S. (2002). Comprehensive expression analysis of time-dependent genetic responses in yeast cells to low temperature. Journal of Biological Chemistry, 277(51), 50015-50021.
BIOL/CMSI 367-01: Biological Databases Fall 2017
Assignments
- Week 1
- Week 2
- Week 3
- Week 4
- Week 5
- Week 6
- Week 7
- Week 8
- Week 9
- Week 10
- Week 11
- Week 12
- Week 14
- Week 15
Journal Entries:
- cwong34 Week 2
- cwong34 Week 3
- cwong34 Week 4
- cwong34 Week 5
- cwong34 Week 6
- cwong34 Week 7
- cwong34 Week 8
- cwong34 Week 9
- cwong34 Week 10
- cwong34 Week 11
- cwong34 Week 12
- cwong34 Week 14
- cwong34 Week 15
Shared Journals:
- cwong34 Week 1 Journal
- cwong34 Week 2 Journal
- cwong34 Week 3 Journal
- cwong34 Week 4 Journal
- cwong34 Week 5 Journal
- cwong34 Week 6 Journal
- cwong34 Week 7 Journal
- cwong34 Week 8 Journal
- cwong34 Week 9 Journal
- cwong34 Week 10 Journal
Group Project