Difference between revisions of "Aporras1 Week 4"
(→Electronic Notebook: final notes on notebook) |
(added zip file) |
||
| (21 intermediate revisions by the same user not shown) | |||
| Line 13: | Line 13: | ||
#*Cyclins are essential because they activate CDKs at the correct times within the cell cycle. | #*Cyclins are essential because they activate CDKs at the correct times within the cell cycle. | ||
#*Provides length of protein to be 546 a.a., PI to be 5.62, and molecular weight to be 62038.2 | #*Provides length of protein to be 546 a.a., PI to be 5.62, and molecular weight to be 62038.2 | ||
| − | #* | + | #*Another observation on the website, it provides additional literature regarding the study of CLN1. |
#I then searched on the NCBI Gene Database and gathered what I thought to be relevant information going from the top to the bottom of the webpage: | #I then searched on the NCBI Gene Database and gathered what I thought to be relevant information going from the top to the bottom of the webpage: | ||
#*A protein coding gene also known as YMR199W | #*A protein coding gene also known as YMR199W | ||
| Line 37: | Line 37: | ||
#*Belongs to the cyclin family. | #*Belongs to the cyclin family. | ||
#*Notices that it cites SGD and must draw from the same if not similar sources. | #*Notices that it cites SGD and must draw from the same if not similar sources. | ||
| + | #Looking at the notes I took here earlier, I created a one paragraph summary of our gene CLN1. | ||
| + | #I then looked at the BioDB for what information was required. | ||
| + | |||
| + | ==One Paragraph Summary on CLN1== | ||
| + | |||
| + | CLN1 is a gene involved in protein coding found on the chromosome XIII. It encodes for a G1 cyclin which regulates the cell cycle meaning they activate CDKs at the correct times within the cell cycle. It can be found within the nucleus and the cytoplasm of the cell. To further characterize CLN1, the genomic sequence is NC_001145.3 and the name of the mRNA and protein Reference Sequences are NM_001182706.1 and NP_013926.1 respectively. Another primary way to identify the gene is through the locus name or tag: YMR199W. The processes CLN1 is involved in includes the cell cycle, cell division, and regulation of serine/threonine kinase activity. Lastly, to discuss the physical characteristics, the protein length was measured to be 546 a.a., the isoelectric point to be 5.62, and the molecular weight to be 62038.2 Da. | ||
| + | |||
| + | ==Additional Information== | ||
| + | |||
| + | '''Standard Name:''' CLN1 | ||
| + | |||
| + | '''Systematic Name:''' YMR199W | ||
| + | |||
| + | '''Name Description:''' cyclin CN1 | ||
| + | |||
| + | '''Gene ID:''' | ||
| + | |||
| + | #'''SGD:''' S000004812 | ||
| + | #*'''Link:'''[[https://www.yeastgenome.org/locus/S000004812 SGD]] | ||
| + | #'''NCBI:''' SGD: S000004812 or Gene ID: 855239 | ||
| + | #*'''Link:'''[[https://www.ncbi.nlm.nih.gov/gene/855239 NCBI]] | ||
| + | #'''Ensembl:''' SGD;Acc: S000004812 | ||
| + | #*'''Link:'''[[https://www.ensembl.org/Saccharomyces_cerevisiae/Transcript/Summary?db=core;g=YMR199W;r=XIII:662644-664284;t=YMR199W Ensembl]] | ||
| + | #'''Unitprot:''' Taxonomic Identifier: 559292 [NCBI] | ||
| + | #*'''Link:'''[[http://www.uniprot.org/uniprot/P20437#subcellular_location Uniprot]] | ||
| + | |||
| + | '''DNA Sequence:''' Chromosome XIII; NC_001145.3 (662644..664284) | ||
| + | |||
| + | 1 ATGAACCACT CAGAAGTGAA AACTGGGTTA ATTGTCACTG CAAAGCAGAC ATATTACCCA | ||
| + | 61 ATTGAATTGT CCAATGCAGA ACTACTAACT CATTACGAAA CCATACAGGA ATATCACGAG | ||
| + | 121 GAAATCTCTC AAAATGTGCT GGTCCAATCT TCCAAGACAA AACCAGACAT AAAATTGATC | ||
| + | 181 GATCAGCAAC CGGAGATGAA TCCTCATCAA ACTAGAGAAG CCATAGTAAC ATTTTTGTAT | ||
| + | 241 CAACTTTCAG TGATGACTAG AGTAAGTAAT GGTATCTTCT TCCACGCTGT CAGGTTCTAC | ||
| + | 301 GATCGCTATT GCTCTAAGAG AGTAGTGTTA AAGGACCAAG CTAAACTAGT TGTAGGCACC | ||
| + | 361 TGCCTTTGGT TAGCGGCCAA AACTTGGGGA GGGTGCAACC ATATTATAAA CAACGTCTCC | ||
| + | 421 ATCCCCACAG GTGGTAGGTT TTATGGTCCC AATCCTAGAG CTCGTATTCC ACGCCTTTCT | ||
| + | 481 GAATTGGTTC ATTATTGCGG CGGGTCCGAT TTATTCGATG AATCAATGTT CATTCAAATG | ||
| + | 541 GAAAGACATA TCTTGGATAC TCTGAACTGG GACGTTTATG AGCCCATGAT TAATGACTAC | ||
| + | 601 ATTTTAAACG TTGACGAAAA TTGTTTGATA CAATATGAAC TTTACAAAAA CCAGTTACAA | ||
| + | 661 AATAACAATA GCAACGGCAA AGAATGGTCC TGTAAGAGAA AGTCACAATC TTCTGACGAC | ||
| + | 721 AGTGATGCCA CAGTGGAAGA ACATATCAGT AGTTCACCGC AAAGTACTGG ACTAGATGGC | ||
| + | 781 GATACAACTA CCATGGATGA AGATGAAGAA CTAAATTCCA AAATTAAATT GATAAATTTG | ||
| + | 841 AAAAGATTCT TAATTGATCT GAGCTGTTGG CAATACAACT TGCTTAAATT CGAATTATAC | ||
| + | 901 GAAATATGCA ATGGTATGTT TTCTATAATA AACAAATTCA CTAATCAAGA TCAAGGCCCT | ||
| + | 961 TTTCTCTCTA TGCCCATTGG TAATGATATA AACTCAAACA CTCAAACGCA GGTATTCAGC | ||
| + | 1021 ATTATCATCA ATGGCATAGT CAATTCTCCC CCATCTTTAG TCGAAGTTTA TAAGGAGCAA | ||
| + | 1081 TATGGTATAG TACCTTTCAT ATTACAAGTA AAAGATTATA ACTTGGAATT ACAAAAGAAA | ||
| + | 1141 CTGCAACTGG CCTCTACAAT AGACCTAACC AGAAAAATTG CTGTCAATTC TCGTTACTTT | ||
| + | 1201 GACCAAAATG CCTCTTCATC ATCAGTTTCT TCTCCAAGCA CATATTCTTC GGGAACCAAT | ||
| + | 1261 TATACTCCAA TGCGAAACTT CAGTGCACAA TCAGACAACA GTGTTTTCAG TACTACCAAC | ||
| + | 1321 ATTGACCATT CATCGCCGAT CACCCCTCAC ATGTACACTT TTAATCAGTT TAAAAACGAA | ||
| + | 1381 AGTGCTTGTG ACAGTGCCAT AAGCGTAAGC AGTCTACCTA ATCAAACCCA AAATGGTAAC | ||
| + | 1441 ATGCCATTAT CAAGCAATTA TCAGAATATG ATGCTAGAAG AAAGGAATAA AGAGAATAGA | ||
| + | 1501 ATTCCCAACT CATCATCCGC TGAAATACCA CAACGTGCCA AATTTATGAC GACTGGTATA | ||
| + | 1561 TTCCAGAACA CAGGAGAACT TACTAATAGA GCGTCTTCGA TATCGCTATC GTTAAGGAAC | ||
| + | 1621 CACAATAGCT CTCAACTGTG A | ||
| + | |||
| + | '''Protein Sequence Corresponding to CLN1:''' NP_013926.1 | ||
| + | |||
| + | 1 MNHSEVKTGL IVTAKQTYYP IELSNAELLT HYETIQEYHE EISQNVLVQS SKTKPDIKLI | ||
| + | 61 DQQPEMNPHQ TREAIVTFLY QLSVMTRVSN GIFFHAVRFY DRYCSKRVVL KDQAKLVVGT | ||
| + | 121 CLWLAAKTWG GCNHIINNVS IPTGGRFYGP NPRARIPRLS ELVHYCGGSD LFDESMFIQM | ||
| + | 181 ERHILDTLNW DVYEPMINDY ILNVDENCLI QYELYKNQLQ NNNSNGKEWS CKRKSQSSDD | ||
| + | 241 SDATVEEHIS SSPQSTGLDG DTTTMDEDEE LNSKIKLINL KRFLIDLSCW QYNLLKFELY | ||
| + | 301 EICNGMFSII NKFTNQDQGP FLSMPIGNDI NSNTQTQVFS IIINGIVNSP PSLVEVYKEQ | ||
| + | 361 YGIVPFILQV KDYNLELQKK LQLASTIDLT RKIAVNSRYF DQNASSSSVS SPSTYSSGTN | ||
| + | 421 YTPMRNFSAQ SDNSVFSTTN IDHSSPITPH MYTFNQFKNE SACDSAISVS SLPNQTQNGN | ||
| + | 481 MPLSSNYQNM MLEERNKENR IPNSSSAEIP QRAKFMTTGI FQNTGELTNR ASSISLSLRN | ||
| + | 541 HNSSQL* | ||
| + | |||
| + | '''Function of CLN1:''' CLN1 Encodes for a G1 cyclin which regulates the cell cycle by activating CDKs at the correct times within the cell cycle thus promoting G1 phase to S phase. | ||
| + | |||
| + | '''Differences about the information provided in each of the parent databases:''' Most of the databases had similar if not overlapping information on CLN1. Most of the databases referenced SGD in particular which went much further in depth including experimental data, shared domains with other genes, and percentage of amino acid in the protein which was particularly interesting. | ||
| + | |||
| + | '''Differences in content, the information, or data itself:''' Most of the overall information was the same regarding data e.g. function, mRNA name, protein name, processes in which CLN1 is involved in and where it's found within the cell. The primary difference was the depth in which each of the sites went into regarding CLN1. | ||
| + | |||
| + | '''Differences in the presentation of the information:''' Most of the sites had easy navigation through the information on CLN1. The only site which was slightly more difficult to navigate was Ensembl because of the setup for the respective sections on the right side of the page. The other three databases had it in one single page and links to each section respectively. | ||
| + | |||
| + | '''Why we chose our gene:''' We chose our gene because we wanted to pick a gene associated with the cell cycle. Specifically, genes that have to do with cancer and mutations in genes that result in cancers e.g. tumor suppressors. Although there weren't tumor suppressor genes for yeast, possible issues with genes promoting the cell cycle was as close as we could go regarding yeast genes. | ||
| + | |||
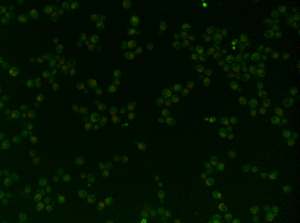
| + | '''Image of yeast strain TKach2012_YMR199W:''' | ||
| + | |||
| + | [[File:Image of YMR199W.png|700px|File:700pixels]] | ||
| + | |||
| + | ==Favorite Gene Page== | ||
| + | |||
| + | [[File:Our Favorite Gene Antonio and Eddie.zip]] | ||
==Acknowledgements== | ==Acknowledgements== | ||
| Line 49: | Line 136: | ||
#Ensembl.org. (2017). Gene: CLN1 (YMR199W) - Supporting evidence - Saccharomyces cerevisiae - Ensembl genome browser 90. [online] Available at: https://www.ensembl.org/Saccharomyces_cerevisiae/Gene/Evidence?db=core;g=YMR199W;r=XIII:662644-664284;t=YMR199W [Accessed 26 Sep. 2017]. | #Ensembl.org. (2017). Gene: CLN1 (YMR199W) - Supporting evidence - Saccharomyces cerevisiae - Ensembl genome browser 90. [online] Available at: https://www.ensembl.org/Saccharomyces_cerevisiae/Gene/Evidence?db=core;g=YMR199W;r=XIII:662644-664284;t=YMR199W [Accessed 26 Sep. 2017]. | ||
| + | # Images.yeastrc.org. (2017). YRC Public Image Repository - Search Results. [online] Available at: http://images.yeastrc.org/imagerepo/searchImageRepo.do?verboseSearchResults=on&dataSource=0&includeFRET=1&channelModifier=3&channelCount=1&zsectionModifier=3&zsectionCount=1×eriesModifier=3×eriesCount=1&objective=0&binning=0&imageSize=0&proteinID=533021&proteinListing=CLN1&proteinID2=0&proteinListing2=&speciesID=0&strainName=&goAcc1=&goAccListing1=&goAcc2=&goAccListing2=&goAcc3=&goAccListing3=&goAcc4=&goAccListing4=&goAcc5=&goAccListing5= [Accessed 26 Sep. 2017]. | ||
#LMU BioDB 2017. (2017). Week 3. Retrieved September 16, 2017, from https://xmlpipedb.cs.lmu.edu/biodb/fall2017/index.php/Week_3 | #LMU BioDB 2017. (2017). Week 3. Retrieved September 16, 2017, from https://xmlpipedb.cs.lmu.edu/biodb/fall2017/index.php/Week_3 | ||
#Ncbi.nlm.nih.gov. (2017). CLN1 cyclin CLN1 [Saccharomyces cerevisiae S288C] - Gene - NCBI. [online] Available at: https://www.ncbi.nlm.nih.gov/gene/855239 [Accessed 26 Sep. 2017]. | #Ncbi.nlm.nih.gov. (2017). CLN1 cyclin CLN1 [Saccharomyces cerevisiae S288C] - Gene - NCBI. [online] Available at: https://www.ncbi.nlm.nih.gov/gene/855239 [Accessed 26 Sep. 2017]. | ||
| + | #Tkach, J. M., Yimit, A., Lee, A. Y., Riffle, M., Costanzo, M., Jaschob, D., ... & Davis, T. N. (2012). Dissecting DNA damage response pathways by analyzing protein localization and abundance changes during DNA replication stress. Nature cell biology, 14(9), 966. | ||
| + | #Uniprot.org. (2017). CLN1 - G1/S-specific cyclin CLN1 - Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) - CLN1 gene & protein. [online] Available at: http://www.uniprot.org/uniprot/P20437#subcellular_location [Accessed 26 Sep. 2017]. | ||
#Yeastgenome.org. (2017). CLN1 | SGD. [online] Available at: https://www.yeastgenome.org/locus/S000004812 [Accessed 26 Sep. 2017]. | #Yeastgenome.org. (2017). CLN1 | SGD. [online] Available at: https://www.yeastgenome.org/locus/S000004812 [Accessed 26 Sep. 2017]. | ||
Latest revision as of 07:02, 26 September 2017
User Page: Antonio Porras
Assignment Page: Week 4
Contents
Electronic Notebook
- I visited the SGD Database and we chose the CLN1 gene as the model organism.
- First, searched through information on the SGD database and found the following going from the top to the bottom of the page:
- Overview: regulation of cell cycle promoting first growth phase to S phase.
- Found on chromosome XIII; also provides the sequence.
- Cyclin dependent protein kinase.
- Found within the nucleus and the cytoplasm.
- Encodes for a G1 cyclin which regulates the cell cycle.
- Cyclins are essential because they activate CDKs at the correct times within the cell cycle.
- Provides length of protein to be 546 a.a., PI to be 5.62, and molecular weight to be 62038.2
- Another observation on the website, it provides additional literature regarding the study of CLN1.
- I then searched on the NCBI Gene Database and gathered what I thought to be relevant information going from the top to the bottom of the webpage:
- A protein coding gene also known as YMR199W
- The genomic sequence is NC_001145.3
- Provides various interactions with other genes, however this list is extensive/exhaustive and not essential to understand the function of the gene.
- Provides further links to PubMed articles on CLN1.
- Ontology processes: cell cycle, cell division, G1/S transition of mitotic cell cycle, regulation of serine/threonine kinase activity.
- Name of the protein: NP_013926
- Name of mRNA: NM_001182706.1
- After exhausting NCBI, I then looked for CLN1 on Ensembl:
- Mostly the same information found on prior websites but I'll include the information anyways.
- Identifier: YMR199W
- Involved in protein coding.
- Found on chromosome 8 (XIII).
- Found in the cytoplasm and the nucleus.
- Involved in processes: cell cycle, cell division, and regulation of cyclin-dependent protein.
- No phenotype associated with the gene.
- Site provides a variant table of the gene, noted to be very extensive.
- Involved in regulation of cell cycle: G1 to S phase.
- As I was looking at the structure of the page, the links to each section were on the left hand side which made it slightly more work to navigate.
- I then searched on uniprot and will only list any information that hasn't yet been mentioned:
- Organism: Saccharomyces cerevisiae (baker's yeast)
- Belongs to the cyclin family.
- Notices that it cites SGD and must draw from the same if not similar sources.
- Looking at the notes I took here earlier, I created a one paragraph summary of our gene CLN1.
- I then looked at the BioDB for what information was required.
One Paragraph Summary on CLN1
CLN1 is a gene involved in protein coding found on the chromosome XIII. It encodes for a G1 cyclin which regulates the cell cycle meaning they activate CDKs at the correct times within the cell cycle. It can be found within the nucleus and the cytoplasm of the cell. To further characterize CLN1, the genomic sequence is NC_001145.3 and the name of the mRNA and protein Reference Sequences are NM_001182706.1 and NP_013926.1 respectively. Another primary way to identify the gene is through the locus name or tag: YMR199W. The processes CLN1 is involved in includes the cell cycle, cell division, and regulation of serine/threonine kinase activity. Lastly, to discuss the physical characteristics, the protein length was measured to be 546 a.a., the isoelectric point to be 5.62, and the molecular weight to be 62038.2 Da.
Additional Information
Standard Name: CLN1
Systematic Name: YMR199W
Name Description: cyclin CN1
Gene ID:
- SGD: S000004812
- Link:[SGD]
- NCBI: SGD: S000004812 or Gene ID: 855239
- Link:[NCBI]
- Ensembl: SGD;Acc: S000004812
- Link:[Ensembl]
- Unitprot: Taxonomic Identifier: 559292 [NCBI]
- Link:[Uniprot]
DNA Sequence: Chromosome XIII; NC_001145.3 (662644..664284)
1 ATGAACCACT CAGAAGTGAA AACTGGGTTA ATTGTCACTG CAAAGCAGAC ATATTACCCA 61 ATTGAATTGT CCAATGCAGA ACTACTAACT CATTACGAAA CCATACAGGA ATATCACGAG 121 GAAATCTCTC AAAATGTGCT GGTCCAATCT TCCAAGACAA AACCAGACAT AAAATTGATC 181 GATCAGCAAC CGGAGATGAA TCCTCATCAA ACTAGAGAAG CCATAGTAAC ATTTTTGTAT 241 CAACTTTCAG TGATGACTAG AGTAAGTAAT GGTATCTTCT TCCACGCTGT CAGGTTCTAC 301 GATCGCTATT GCTCTAAGAG AGTAGTGTTA AAGGACCAAG CTAAACTAGT TGTAGGCACC 361 TGCCTTTGGT TAGCGGCCAA AACTTGGGGA GGGTGCAACC ATATTATAAA CAACGTCTCC 421 ATCCCCACAG GTGGTAGGTT TTATGGTCCC AATCCTAGAG CTCGTATTCC ACGCCTTTCT 481 GAATTGGTTC ATTATTGCGG CGGGTCCGAT TTATTCGATG AATCAATGTT CATTCAAATG 541 GAAAGACATA TCTTGGATAC TCTGAACTGG GACGTTTATG AGCCCATGAT TAATGACTAC 601 ATTTTAAACG TTGACGAAAA TTGTTTGATA CAATATGAAC TTTACAAAAA CCAGTTACAA 661 AATAACAATA GCAACGGCAA AGAATGGTCC TGTAAGAGAA AGTCACAATC TTCTGACGAC 721 AGTGATGCCA CAGTGGAAGA ACATATCAGT AGTTCACCGC AAAGTACTGG ACTAGATGGC 781 GATACAACTA CCATGGATGA AGATGAAGAA CTAAATTCCA AAATTAAATT GATAAATTTG 841 AAAAGATTCT TAATTGATCT GAGCTGTTGG CAATACAACT TGCTTAAATT CGAATTATAC 901 GAAATATGCA ATGGTATGTT TTCTATAATA AACAAATTCA CTAATCAAGA TCAAGGCCCT 961 TTTCTCTCTA TGCCCATTGG TAATGATATA AACTCAAACA CTCAAACGCA GGTATTCAGC 1021 ATTATCATCA ATGGCATAGT CAATTCTCCC CCATCTTTAG TCGAAGTTTA TAAGGAGCAA 1081 TATGGTATAG TACCTTTCAT ATTACAAGTA AAAGATTATA ACTTGGAATT ACAAAAGAAA 1141 CTGCAACTGG CCTCTACAAT AGACCTAACC AGAAAAATTG CTGTCAATTC TCGTTACTTT 1201 GACCAAAATG CCTCTTCATC ATCAGTTTCT TCTCCAAGCA CATATTCTTC GGGAACCAAT 1261 TATACTCCAA TGCGAAACTT CAGTGCACAA TCAGACAACA GTGTTTTCAG TACTACCAAC 1321 ATTGACCATT CATCGCCGAT CACCCCTCAC ATGTACACTT TTAATCAGTT TAAAAACGAA 1381 AGTGCTTGTG ACAGTGCCAT AAGCGTAAGC AGTCTACCTA ATCAAACCCA AAATGGTAAC 1441 ATGCCATTAT CAAGCAATTA TCAGAATATG ATGCTAGAAG AAAGGAATAA AGAGAATAGA 1501 ATTCCCAACT CATCATCCGC TGAAATACCA CAACGTGCCA AATTTATGAC GACTGGTATA 1561 TTCCAGAACA CAGGAGAACT TACTAATAGA GCGTCTTCGA TATCGCTATC GTTAAGGAAC 1621 CACAATAGCT CTCAACTGTG A
Protein Sequence Corresponding to CLN1: NP_013926.1
1 MNHSEVKTGL IVTAKQTYYP IELSNAELLT HYETIQEYHE EISQNVLVQS SKTKPDIKLI 61 DQQPEMNPHQ TREAIVTFLY QLSVMTRVSN GIFFHAVRFY DRYCSKRVVL KDQAKLVVGT 121 CLWLAAKTWG GCNHIINNVS IPTGGRFYGP NPRARIPRLS ELVHYCGGSD LFDESMFIQM 181 ERHILDTLNW DVYEPMINDY ILNVDENCLI QYELYKNQLQ NNNSNGKEWS CKRKSQSSDD 241 SDATVEEHIS SSPQSTGLDG DTTTMDEDEE LNSKIKLINL KRFLIDLSCW QYNLLKFELY 301 EICNGMFSII NKFTNQDQGP FLSMPIGNDI NSNTQTQVFS IIINGIVNSP PSLVEVYKEQ 361 YGIVPFILQV KDYNLELQKK LQLASTIDLT RKIAVNSRYF DQNASSSSVS SPSTYSSGTN 421 YTPMRNFSAQ SDNSVFSTTN IDHSSPITPH MYTFNQFKNE SACDSAISVS SLPNQTQNGN 481 MPLSSNYQNM MLEERNKENR IPNSSSAEIP QRAKFMTTGI FQNTGELTNR ASSISLSLRN 541 HNSSQL*
Function of CLN1: CLN1 Encodes for a G1 cyclin which regulates the cell cycle by activating CDKs at the correct times within the cell cycle thus promoting G1 phase to S phase.
Differences about the information provided in each of the parent databases: Most of the databases had similar if not overlapping information on CLN1. Most of the databases referenced SGD in particular which went much further in depth including experimental data, shared domains with other genes, and percentage of amino acid in the protein which was particularly interesting.
Differences in content, the information, or data itself: Most of the overall information was the same regarding data e.g. function, mRNA name, protein name, processes in which CLN1 is involved in and where it's found within the cell. The primary difference was the depth in which each of the sites went into regarding CLN1.
Differences in the presentation of the information: Most of the sites had easy navigation through the information on CLN1. The only site which was slightly more difficult to navigate was Ensembl because of the setup for the respective sections on the right side of the page. The other three databases had it in one single page and links to each section respectively.
Why we chose our gene: We chose our gene because we wanted to pick a gene associated with the cell cycle. Specifically, genes that have to do with cancer and mutations in genes that result in cancers e.g. tumor suppressors. Although there weren't tumor suppressor genes for yeast, possible issues with genes promoting the cell cycle was as close as we could go regarding yeast genes.
Image of yeast strain TKach2012_YMR199W:
Favorite Gene Page
File:Our Favorite Gene Antonio and Eddie.zip
Acknowledgements
- Met outside of class with Eddie Azinge to discuss any questions we had prior to meeting and throughout the process of completing the Week 4 assignment.
While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source.
Aporras1 (talk) 21:03, 25 September 2017 (PDT)
References
- Ensembl.org. (2017). Gene: CLN1 (YMR199W) - Supporting evidence - Saccharomyces cerevisiae - Ensembl genome browser 90. [online] Available at: https://www.ensembl.org/Saccharomyces_cerevisiae/Gene/Evidence?db=core;g=YMR199W;r=XIII:662644-664284;t=YMR199W [Accessed 26 Sep. 2017].
- Images.yeastrc.org. (2017). YRC Public Image Repository - Search Results. [online] Available at: http://images.yeastrc.org/imagerepo/searchImageRepo.do?verboseSearchResults=on&dataSource=0&includeFRET=1&channelModifier=3&channelCount=1&zsectionModifier=3&zsectionCount=1×eriesModifier=3×eriesCount=1&objective=0&binning=0&imageSize=0&proteinID=533021&proteinListing=CLN1&proteinID2=0&proteinListing2=&speciesID=0&strainName=&goAcc1=&goAccListing1=&goAcc2=&goAccListing2=&goAcc3=&goAccListing3=&goAcc4=&goAccListing4=&goAcc5=&goAccListing5= [Accessed 26 Sep. 2017].
- LMU BioDB 2017. (2017). Week 3. Retrieved September 16, 2017, from https://xmlpipedb.cs.lmu.edu/biodb/fall2017/index.php/Week_3
- Ncbi.nlm.nih.gov. (2017). CLN1 cyclin CLN1 [Saccharomyces cerevisiae S288C] - Gene - NCBI. [online] Available at: https://www.ncbi.nlm.nih.gov/gene/855239 [Accessed 26 Sep. 2017].
- Tkach, J. M., Yimit, A., Lee, A. Y., Riffle, M., Costanzo, M., Jaschob, D., ... & Davis, T. N. (2012). Dissecting DNA damage response pathways by analyzing protein localization and abundance changes during DNA replication stress. Nature cell biology, 14(9), 966.
- Uniprot.org. (2017). CLN1 - G1/S-specific cyclin CLN1 - Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) - CLN1 gene & protein. [online] Available at: http://www.uniprot.org/uniprot/P20437#subcellular_location [Accessed 26 Sep. 2017].
- Yeastgenome.org. (2017). CLN1 | SGD. [online] Available at: https://www.yeastgenome.org/locus/S000004812 [Accessed 26 Sep. 2017].