Difference between revisions of "QLanners Week 14"
(added rest of answers for hsf1 gene) |
(added references an titles of other sections) |
||
| Line 1: | Line 1: | ||
| − | + | ==Electronic Journal== | |
| + | In order to determine what fields should be taken from each database, I worked closely with [[User:Cwong34|Corinne Wong]] and [[User:Kwrigh35|Katie Wright]]. We the gene HSF1, which is a transcription factor, to determine which fields should be pulled from each database. All of the fields are included in the below section as a bullet. Each bullet is followed by a portion of italicized text which corresponds to the information of that field for HSF1. | ||
| + | ===Brainstorming=== | ||
General info we want about each gene: | General info we want about each gene: | ||
*Gene ID from each database | *Gene ID from each database | ||
| Line 14: | Line 16: | ||
*Gene Ontology (SGD - see if we can find it on UniProt) | *Gene Ontology (SGD - see if we can find it on UniProt) | ||
| + | ===Fields to Pull From Each Database=== | ||
We decided that from JASPAR we will pull: | We decided that from JASPAR we will pull: | ||
*Matrix ID ''MA0319.1'' | *Matrix ID ''MA0319.1'' | ||
| Line 98: | Line 101: | ||
***''Manually Curated: nucleus (IDA)'' | ***''Manually Curated: nucleus (IDA)'' | ||
***''High-Throughput: mitochondrion (HDA)'' | ***''High-Throughput: mitochondrion (HDA)'' | ||
| + | |||
| + | ===Other Tasks=== | ||
| + | Along with working on determining what | ||
| + | |||
| + | ==Acknowledgements== | ||
| + | |||
| + | ==References== | ||
| + | Ensembl. (2017). "Gene: HSF1". Retrieved November 28, 2017, from https://www.ensembl.org/Saccharomyces_cerevisiae/Gene/Summary?db=core;g=YGL073W;r=VII:368753-371254;t=YGL073W <br> | ||
| + | JASPAR. (2017). "Detailed information of matrix profile MA0319.1". Retrieved on November 28, 2017, from http://jaspar.genereg.net/matrix/MA0319.1/ <br> | ||
| + | LMU BioDB 2017. (2017). Week 14. Retrieved November 28, 2017, from https://xmlpipedb.cs.lmu.edu/biodb/fall2017/index.php/Week_14 <br> | ||
| + | NCBI. (2017). "HSF1 stress-responsive transcription factor HSF1 [ Saccharomyces cerevisiae S288C ]". Retrieved November 28, 2017, from https://www.ncbi.nlm.nih.gov/gene/852806 <br> | ||
| + | Saccharomyces Genome Database. (2017). "HSF1 / YGL073W Overview". Retrieved November 28, 2017, from https://www.yeastgenome.org/locus/S000003041 <br> | ||
| + | UniProt. (2017). "UniProtKB - P10961 (HSF_YEAST)". Retrieved November 28, 2017 from http://www.uniprot.org/uniprot/P10961 | ||
Revision as of 04:45, 5 December 2017
Contents
Electronic Journal
In order to determine what fields should be taken from each database, I worked closely with Corinne Wong and Katie Wright. We the gene HSF1, which is a transcription factor, to determine which fields should be pulled from each database. All of the fields are included in the below section as a bullet. Each bullet is followed by a portion of italicized text which corresponds to the information of that field for HSF1.
Brainstorming
General info we want about each gene:
- Gene ID from each database
- Description/Function (ensembl)
- DNA Sequence (ensembl)
- Protein Sequence (UniProt)
- Locus tag (NCBI)
- Also Known As (NCBI)
- Consensus Sequence (JASPAR)
- Regulation (SGD)
- Interaction (SGD)
- Similar Proteins (UniProt)
- Gene Ontology (SGD - see if we can find it on UniProt)
Fields to Pull From Each Database
We decided that from JASPAR we will pull:
- Matrix ID MA0319.1
- Class Heat shock factors
- Family HSF factors
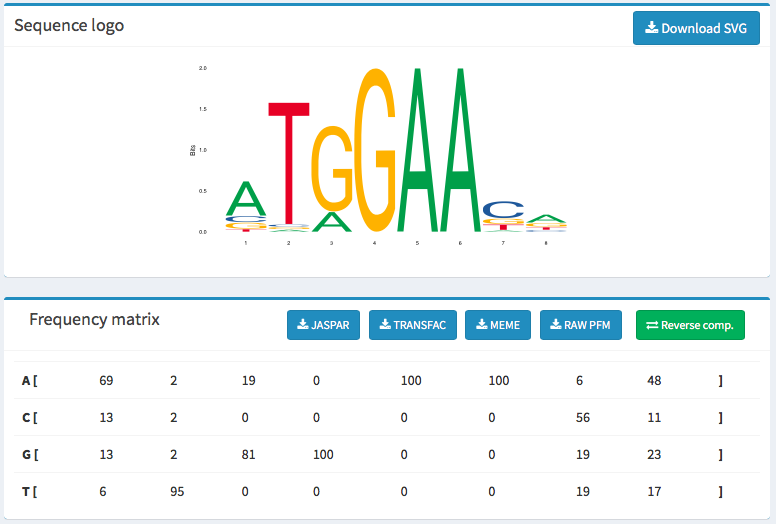
- Sequence Logo image below
- Frequency Matrix image below
Breakdown of what we want from all other databases:
NCBI:
- Gene ID 852806
- Locus Tag YGL073W
- Also Known As EXA3; MAS3
- Chromosome Sequence Chromosome: VII; NC_001139.9 (368753..371254)
- Genomic Sequence NC_001139.9
- Protein Sequence NP_011442.3
Ensembl:
- Gene ID YGL073W
- Description/Function Trimeric heat shock transcription factor; activates multiple genes in response to highly diverse stresses, including hyperthermia; recognizes variable heat shock elements (HSEs) consisting of inverted NGAAN repeats; monitors translational status of cell at the ribosome through an RQC (Ribosomal Quality Control)-mediated translation-stress signal; involved in diauxic shift; posttranslationally regulated [Source:SGD;Acc:S000003041]
- DNA Sequence >chromosome:R64-1-1:VII:368153:371854:1
AAAATACTCCACTAAGGCCAGTAGCAACAACACGTTTTCTTGGATGATGCGTTTTCTTGA ACAAACAGTACCGACTAGGACTGTTTCAATGAAGTTGTGTACGGTCTGGTAGTATATCTA TATTCCGTGATGCCTTTGTGGAGGACGTTGAGATGAGACTGAGTCGTACACCATGTTATT CCTGTTTACGGTTAATTGCGCGTCGCGCTTTCTCTAGCAAATATCTCGGTTCGAAGTAAA GCAGGTCCTTCATGTAATGGTAACCTAAGGCAAAGGGTTTGTCATATACCCGTGAAGGCA TTTACACAAGCGCACTTCTAGTCATATGCAGTTCATGCATATTAAGTGAGTGTTATAACG CAAGAGTTATATTTGAAATAGGGTTGTTAAAGAAGGGAGAACCCATTCACCACATTATCT TTGCGAGTGTAAAACTAGATAACTTAAATTTTTAGGAGAGATTTTGCCACTTGGCAGCAA ATACCAAATAGCAGTACTGTTCCGGTAGATAAAGGCAAAGAGTTAGAGGTGTGCTTTACG AACAGCGCTGGAAGGGAAAGGAAACAAAAAAGACAAAAAGACAGCTGTATTGTTGGCGCC ATGAATAATGCTGCAAATACAGGGACGACCAATGAGTCAAACGTGAGCGATGCTCCCCGT ATTGAGCCTTTACCAAGCTTGAATGATGATGACATTGAAAAAATCTTACAACCGAACGAT ATCTTTACGACCGATCGTACCGATGCAAGTACTACATCTTCCACAGCCATTGAAGATATT ATTAACCCCTCATTGGATCCGCAGTCAGCAGCATCGCCGGTTCCTTCTTCCTCTTTTTTC CATGACTCAAGGAAACCTTCCACCAGTACACATTTAGTAAGGAGAGGTACTCCATTGGGA ATTTACCAAACCAATCTATACGGTCACAATAGCAGAGAAAATACTAATCCTAATAGTACA TTATTATCTTCTAAGTTACTCGCGCATCCACCAGTTCCTTATGGGCAAAATCCCGATTTA CTACAACATGCTGTGTACAGGGCACAGCCGTCAAGTGGAACCACTAACGCGCAACCGCGC CAAACCACAAGAAGATATCAATCCCATAAATCACGGCCTGCATTTGTTAATAAACTATGG AGCATGTTAAACGATGATTCTAATACGAAACTTATACAGTGGGCGGAGGATGGAAAATCT TTTATTGTCACGAATAGGGAGGAATTTGTGCACCAAATTTTACCAAAATATTTTAAACAT TCCAATTTCGCTTCCTTTGTAAGACAATTGAACATGTATGGATGGCATAAAGTTCAAGAT GTCAAGTCAGGATCAATTCAAAGTAGTTCAGATGATAAGTGGCAATTTGAAAATGAAAAC TTCATTAGAGGTAGAGAAGATTTGCTGGAAAAAATAATCAGGCAGAAAGGTTCCTCCAAT AACCATAATAGCCCTAGTGGTAACGGTAATCCAGCGAATGGTAGCAACATCCCTCTGGAC AATGCCGCAGGAAGTAATAATAGCAATAATAACATCAGTAGTAGTAATTCATTTTTTAAC AATGGTCATTTATTGCAGGGTAAAACACTAAGATTAATGAACGAAGCGAATCTTGGAGAT AAGAATGATGTCACCGCGATTTTGGGGGAATTAGAGCAAATAAAATATAACCAGATTGCA ATTTCCAAAGATTTACTAAGAATAAACAAAGATAATGAGTTATTATGGCAAGAGAATATG ATGGCCAGGGAAAGACATAGAACCCAACAGCAAGCCTTGGAAAAAATGTTCAGATTCTTG ACATCTATAGTCCCACACTTAGATCCCAAAATGATTATGGACGGGCTGGGAGATCCGAAA GTTAATAATGAAAAGCTAAACAGTGCGAATAACATTGGGTTAAATCGCGACAACACAGGC ACTATAGATGAACTAAAATCCAACGATTCTTTCATAAACGATGATCGTAATTCTTTCACC AATGCTACAACCAACGCCCGTAATAACATGAGTCCCAACAATGATGACAATAGTATTGAC ACCGCTAGCACTAATACCACCAACAGAAAGAAAAATATAGATGAAAACATCAAAAATAAC AACGACATAATTAATGACATTATATTTAATACCAACCTTGCCAACAATCTCAGCAATTAC AATTCCAACAATAATGCTGGCTCGCCAATAAGGCCCTATAAACAAAGATATCTTTTGAAA AATAGAGCCAATTCCTCGACATCGAGTGAGAATCCAAGCCTAACGCCCTTTGATATCGAA TCTAATAATGACCGCAAAATTTCAGAAATTCCTTTTGATGACGAAGAAGAAGAAGAAACG GATTTTAGGCCTTTTACCTCGCGAGATCCTAATAACCAAACGAGTGAAAACACTTTTGAT CCAAACAGATTTACGATGCTCTCTGATGATGATTTAAAAAAAGATTCTCATACCAATGAC AATAAACACAACGAAAGTGATCTTTTTTGGGACAACGTACATAGAAATATAGACGAACAA GATGCAAGACTCCAGAACTTGGAAAATATGGTTCACATACTTTCTCCTGGATATCCTAAT AAGTCGTTCAACAACAAAACTTCCTCGACAAACACTAATTCCAATATGGAAAGTGCTGTC AACGTTAATAGCCCTGGTTTCAACTTACAGGATTATTTAACTGGAGAGTCTAATTCCCCC AATTCTGTTCATTCTGTTCCCTCCAATGGCAGCGGCTCCACACCGTTGCCCATGCCAAAT GATAATGACACCGAGCACGCAAGTACAAGTGTCAATCAAGGCGAAAATGGAAGCGGATTA ACGCCCTTCCTCACGGTAGATGATCACACACTAAACGACAATAACACTAGTGAGGGAAGT ACAAGGGTGTCCCCCGATATAAAGTTCAGCGCCACTGAAAACACTAAAGTGAGTGATAAC CTGCCAAGCTTTAATGACCACAGTTATTCCACCCAGGCCGACACGGCGCCCGAGAACGCT AAGAAAAGATTTGTGGAGGAAATACCGGAACCGGCTATAGTCGAAATACAGGACCCGACA GAGTACAACGATCACCGCCTGCCCAAACGAGCTAAGAAATAGTACACAGGGCAAGGTCAT TAAATAGCGTATATAATCATTTAATATAGTATGTTCTCGAAGCTGATCGCGTAAGGCGCA GAGCGAACTAAAAAAAATACCGGCACCCATGCACCTCACACCGCCGCACGCGAGTGAGGT TGAACTGCACCCGGAAAATGCCAAGTAGATGAGTCGTGAAGAGTTCTCGTTATTCGAGCT AGTGAGAGCCTGAGAAGGGCTTGCCGAGTGAACTGGTGTCACATTGGCCGTTTTAACGCA AGTTGGCGTACTTATATTGACTGTTGGATGAAAGGGTAATCAAGAGAAACGGAAACGGCC TCCTCATCGTTAAGCTCATCAGTATTCATTTCTCCCCTTTCTGCTCCATCGCGTGCTCGA GACTATATTCTTCAGATTATCAAGCAGAAACAGAATTCGCATATTACATAACTTTCACAG GTTGAAGTATAAACCGCTACAGTACACAACCTCGGATAGAATATAGGGAAGAGGCCAATT CCGTGAAAACGATTTAATATTCTTTACAGTTACAAAAAGTATTACCTATTATCCTCTTTT CGGTGTCATTGACAAACCTCTTAGCGACAGAAACTCCCTAGC
- Gene Location Chromosome VII: 368,753-371,254
- 'Gene Map
[File:Saccharomycescerevisiae HSF1.pdf]
UniProt:
- Gene ID: P10961 (HSF_YEAST)
- Protein Sequence
MNNAANTGTTNESNVSDAPRIEPLPSLNDDDIEKILQPNDIFTTDRTDASTTSSTAIEDI
INPSLDPQSAASPVPSSSFFHDSRKPSTSTHLVRRGTPLGIYQTNLYGHNSRENTNPNST
LLSSKLLAHPPVPYGQNPDLLQHAVYRAQPSSGTTNAQPRQTTRRYQSHKSRPAFVNKLW
SMLNDDSNTKLIQWAEDGKSFIVTNREEFVHQILPKYFKHSNFASFVRQLNMYGWHKVQD
VKSGSIQSSSDDKWQFENENFIRGREDLLEKIIRQKGSSNNHNSPSGNGNPANGSNIPLD
NAAGSNNSNNNISSSNSFFNNGHLLQGKTLRLMNEANLGDKNDVTAILGELEQIKYNQIA
ISKDLLRINKDNELLWQENMMARERHRTQQQALEKMFRFLTSIVPHLDPKMIMDGLGDPK
VNNEKLNSANNIGLNRDNTGTIDELKSNDSFINDDRNSFTNATTNARNNMSPNNDDNSID
TASTNTTNRKKNIDENIKNNNDIINDIIFNTNLANNLSNYNSNNNAGSPIRPYKQRYLLK
NRANSSTSSENPSLTPFDIESNNDRKISEIPFDDEEEEETDFRPFTSRDPNNQTSENTFD
PNRFTMLSDDDLKKDSHTNDNKHNESDLFWDNVHRNIDEQDARLQNLENMVHILSPGYPN
KSFNNKTSSTNTNSNMESAVNVNSPGFNLQDYLTGESNSPNSVHSVPSNGSGSTPLPMPN
DNDTEHASTSVNQGENGSGLTPFLTVDDHTLNDNNTSEGSTRVSPDIKFSATENTKVSDN
LPSFNDHSYSTQADTAPENAKKRFVEEIPEPAIVEIQDPTEYNDHRLPKRAKK
- Similar Protein: N1P1W2 and ID of Similar Protein: P10961
- Protein Type/Name: Heat shock factor protein
- Species: Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast)
SGD:
- Gene ID
- Standard Name HSF1
- Systematic Name YGL073W
- SGD ID S000003041
- Regulation
- Regulators: 6
- Targets: 478
- Interaction
- Total Interactions: 85 total interactions for 71 unique genes
- Physical Interactions:
- Affinity Capture-MS: 11
- Affinity Capture-RNA: 1
- Affinity Capture-Western: 4
- Biochemical Activity: 11
- Co-localization: 3
- Reconstituted Complex: 2
- Two-hybrid: 3
- Genetic Interactions:
- Dosage Rescue: 16
- Negative Genetic: 8
- Phenotypic Enhancement: 1
- Phenotypic Suppression: 5
- Synthetic Growth Defect: 2
- Synthetic Haploinsufficiency: 1
- Synthetic Lethality: 6
- Synthetic Rescue: 11
- Gene Ontology
- Summary: Sequence-specific DNA binding transcription factor that induces expression of the Hsp90-family protein chaperones Hsc82p and Hsp82p during the cellular response to heat; also negatively regulates TOR signaling
- Molecular Function:
- Manually Curated: DNA binding transcription factor activity (IDA)
- High-Throughput: sequence-specific DNA binding (HDA)
- Biological Process
- Manually Curated: negative regulation of TOR signaling (IMP), positive regulation of transcription from RNA polymerase II promoter (IMP), regulation of establishment of protein localization to chromosome (IMP), regulation of transcription from RNA polymerase II promoter (IDA), response to heat (IMP)
- Cellular Component:
- Manually Curated: nucleus (IDA)
- High-Throughput: mitochondrion (HDA)
Other Tasks
Along with working on determining what
Acknowledgements
References
Ensembl. (2017). "Gene: HSF1". Retrieved November 28, 2017, from https://www.ensembl.org/Saccharomyces_cerevisiae/Gene/Summary?db=core;g=YGL073W;r=VII:368753-371254;t=YGL073W
JASPAR. (2017). "Detailed information of matrix profile MA0319.1". Retrieved on November 28, 2017, from http://jaspar.genereg.net/matrix/MA0319.1/
LMU BioDB 2017. (2017). Week 14. Retrieved November 28, 2017, from https://xmlpipedb.cs.lmu.edu/biodb/fall2017/index.php/Week_14
NCBI. (2017). "HSF1 stress-responsive transcription factor HSF1 [ Saccharomyces cerevisiae S288C ]". Retrieved November 28, 2017, from https://www.ncbi.nlm.nih.gov/gene/852806
Saccharomyces Genome Database. (2017). "HSF1 / YGL073W Overview". Retrieved November 28, 2017, from https://www.yeastgenome.org/locus/S000003041
UniProt. (2017). "UniProtKB - P10961 (HSF_YEAST)". Retrieved November 28, 2017 from http://www.uniprot.org/uniprot/P10961