Kwrigh35 Week 3
Contents
Hack a Page
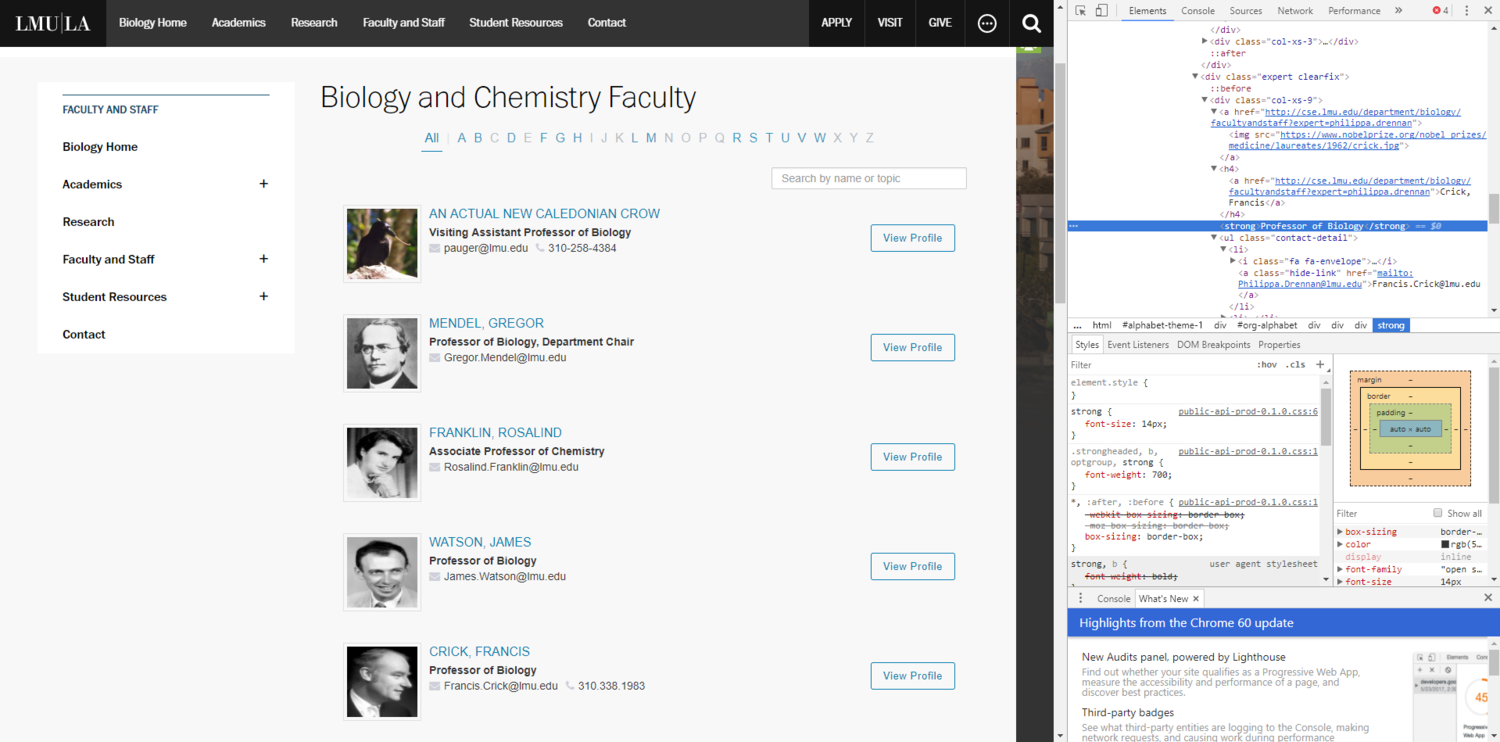
Screenshot with Developer Tools Open
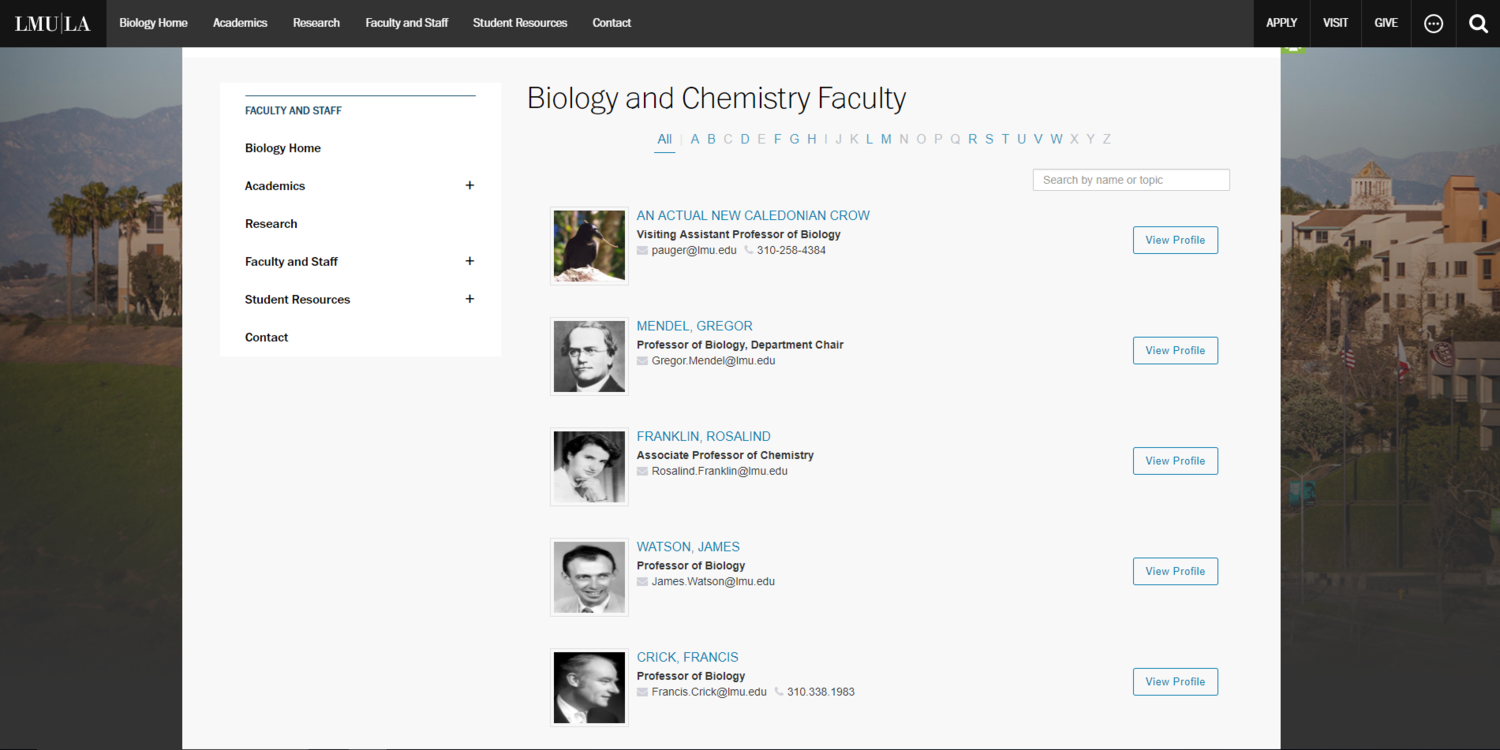
Screenshot without Developer Tools Open
The Genetic Code, by Way of the Web
Curl Command
Here is the curl command that requests a translation at the command-line, raw-data level
curl -d "pre_text=cgatggtacatggagtccagtagccgtagtgatgagatcgatgagctagc&output=Verbose ("Metp", spaces between residues) &code=Standard" http://web.expasy.org/cgi-bin/translate/dna_aa
The result of this command was compared with the HTML from the ExPASy page and the reading frames were confirmed to be the same.
Study the Curl'd Code
Question 1
I found 3 links to other pages on the web.expasy.org server. I also found a link to the file I had saved the curl'd html to. However, since I was viewing an .html file on my computer, the server it tried to access was on the local computer and not the ExPASy server. Therefore, it was represented as a file path and not a webpage.
file:///C:/Users/biolab/Desktop/Week%203%20practice.html (the file itself)
file:///C:/css/sib_css/sib.css
file:///C:/css/base.css
file:///C:/css/sib_css/sib_print.css
I found another link to the Google-Analytics server. This link was present twice, for a grand total of six requests.
http://www.google-analytics.com/ga.js
Using the command line to extract just the answers
This command will just give six reading frames from the DNA sequence provided. See Notebook section for process.
curl -d "pre_text=cgatggtacatggagtccagtagccgtagtgatgagatcgatgagctagc&output=Verbose ("Met", "Stop", spaces between residues) &code=Standard" http://web.expasy.org/cgi-bin/translate/dna_aa | sed "1,47d" | sed "14,44d" | sed 's/<[^>]*>//g' | sed "2s/[A-Z]/& /g" | sed "4s/[A-Z]/& /g" | sed "6s/[A-Z]/& /g" | sed "8s/[A-Z]/& /g" | sed "10s/[A-Z]/& /g" | sed "12s/[A-Z]/& /g" | sed "s/M/Met/g"| sed "s/-/Stop /g"
Notebook
Hack-a-Page
For this portion of the assignment, I decided that I wanted LMU to hire some famous new faculty members. Most of the new faculty members are famous scientists who have made major contributions to the field of Biology. I also wanted LMU to hire a New Caledonian Crow, because Professor Auger insists that they are one of the most intelligent animal species on Earth. I found the images by doing a simple google search for each of them. To replace them, I opened developer tools, used the element selector tool (Ctrl+Shift+C) to click on the image I wanted to select, and then pasted the image url directly into the html. I also used the selector tool to find the html for the professor's names and emails and replaced those.
The Genetic Code, by Way of the Web
- My homework partner John and I worked on this portion of the assignment together on Monday 9/18 and struggled a lot. We each worked semi independently, but talked about our ideas and tried different manipulations of the code.
- 9/19: To figure out the curl command, I consulted with John's week 3 journal page. He showed the curl command as a formula, so to confirm that I knew how to use this "formula," I added the appropriate values for pre_text, output, and code. The HTML that Ubuntu gave back to me included the same amino acid sequences/reading frames that I got back from ExPASy when I submitted the DNA sequence with the same parameters. This indicates that I had found the correct curl command.
- 9/19: To find the links, I went to my web browser and Initially I studied the requests that the ExPASy site made itself. I just realized that I should be looking at the requests that my curled code made instead, so I am going to go back and amend my answers right now. I plan on opening the .html file that I saved (containing the results I got from my curl'd command) with chrome and using the developer tools to look at the requests made.
- 9/19: To extract just the answers: I used the curl command that I discussed earlier, but I was having trouble figuring out how to use sed to delete/select multiple lines of code. I went to Zach's Week 3 journal to figure out how he did it, then just copy and pasted his sed commands after my curl code.
Acknowledgements
- I would like to thank my homework partner John Lopez for his help on this assignment. We worked together Monday afternoon on "The Genetic Code, by Way of the Web" part of the assignment. We struggled to complete this portion of the assignment, and since it was extended, we decided to ask Professor Dionisio for help after class on Tuesday afternoon. Kwrigh35 (talk) 16:27, 19 September 2017 (PDT)
- I also looked at John's week 3 assignment page to help me Tuesday evening, since he had reviewed some material with Professor Dionisio.
- I'd like to thank Professor Dionisio for assisting my homework partner, who in turn assisted me.
- Thanks to Zach as well. I looked at his journal entry for help on the "extract just the answers" section and copied part of his code.
- While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source. Kwrigh35 (talk) 22:51, 19 September 2017 (PDT)
References
LMU BioDB 2017. (2017). Week 3. Retrieved September 18, 2017, from Week 3
LMU BioDB 2017. (2017). Dynamic Text Processing. Retrieved September 18, 2017, from Dynamic Text Processing
LMU BioDB 2017. (2017). Introduction to the Command Line. Retrieved September 18, 2017, from Introduction to the Command Line
Useful Links
| Category | Links |
|---|---|
| Interwiki | Main Page Katie Wright Course schedule Lights, Camera, InterACTION! |
| Katie Wright Weekly Assignment |
Week 2 Week 3 Week 4 Week 5 Week 6 Week 7 Week 8 Week 9🎃 Week 10 Week 11 Week 12 Week 13 Week 14 Week 15 |
| Weekly Assignment Instructions |
Week 1 Week 2 Week 3 Week 4 Week 5 Week 6 Week 7 Week 8 Week 9 Week 10 Week 11 Week 12 Week 14 Week 15 |
| Class Journals | Week 1 Week 2 Week 3 Week 4 Week 5 Week 6 Week 7 Week 8 Week 9 Week 10 Week 11 Week 12 Week 13 Week 14 |