Simonwro120 Week 3
From LMU BioDB 2017
Contents
Electronic Laboratory Notebook
Hack-a-Page
With DevTools
Without DevTools
The Genetic Code, by Way of the Web
curl Command
curl -d "pre_text=cgtatgctaataccatgttccgcgtataacccagccgccagttccgctggcggcatttta&submit=TRANSLATE SEQUENCE" http://web.expasy.org/cgi-bin/translate/dna_aa
ExPASy Questions
Question 1
The server's response does have some links to other pages which include:
- This is a link to a page full of text resembling code and titled Swiss Institute of Bioinformatics.
- This is a link to an their logo.
- This link is very similar to my first. Just another text page titled Swiss Institute of Bioinformatics.
- This is a link to a wikipedia page explaining Open reading frames as they pertain to biology.
- This link is similar to 1 and 3. Another text file, however this one is titles CSS for Genevian Resources.
Question 2
There are identifiers in the ExPASy translation server’s responses which include:
- "sib_footer" identifier pointing to the bottom of the page.
- "sib_headrer" identifier pointing towards the top of the page.
- "pre_text" identifier naming the box where text may go.
- "POST" identifier tells the browser how to contact the server.
Just the Answers Using the Command Line
curl "http://web.expasy.org/cgi-bin/translate/dna_aa?pre_text=cgatggtacatggagtccagtagccgtagtgatgagatcgatgagctagc&output=Verbose&code=Standard" | grep -E '<(BR|PRE)>' | sed 's/<[^>]*>//g'
Electronic Lab Notebook
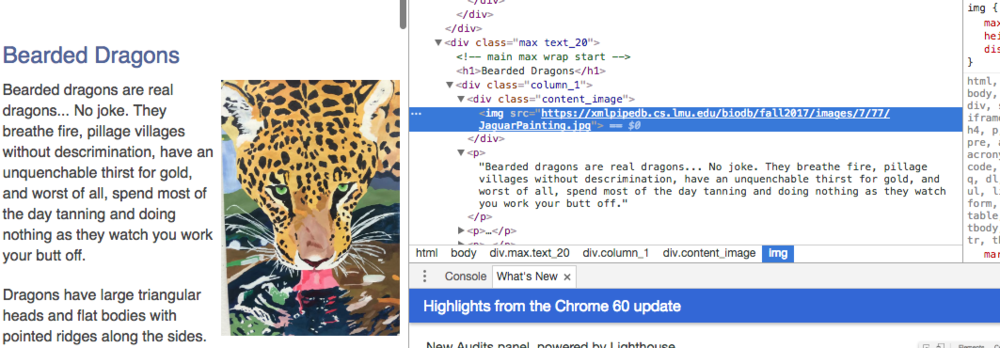
For Hack-a-Page:
- First I accessed a website of my choosing and right clicked in Google Chrome to induce the dropdown menu.
- I then clicked on "Inspect" to bring up the developer tools.
- I then found a paragraph located next to a linked image and chose that part of the website as my target for modification.
- After that, all I had to do was find the right code, modify it in any way I saw fit, and take my screenshots.
For "curl" Commands:
- First I looked over a few basic curl commands to get a feel for how it works.
- I then searched for commands that would allow for user input, as well ass a command that would allow me to activate a button most likely associated to some kind of script.
- After some research, I started to fiddle wit the command line.
- To verify that my command was correct, I searched the the code that was returned by the terminal representing the amino acid chains and cross referenced them with the already known correct sequences.
List of Assignments
Week 1 Week 2 Week 3 Week 4 Week 5 Week 6 Week 7 Week 8 Week 9 Week 10 Week 11 Week 12 Week 13 Week 14 Week 15
List of Journal Entries
Week 1 Week 2 Week 3 Week 4 Week 5 Week 6 Week 7 Week 8 Week 9 Week 10 Week 11 Week 12 Week 13 Week 14 Week 15
Week 1 Week 2 Week 3 Week 4 Week 5 Week 6 Week 7 Week 8 Week 9 Week 10 Week 11 Week 12 Week 13 Week 14 Week 15
Acknowledgements
- Thanks to Doctors Dahquist and Dondi to helping clarify the assignments instructions.
- Thanks to "subfuzion' an account name for someone who helped me on gitHub.
- Special thanks to Eddie Azinge and Blair Hamilton. With their help, I was able to learn what the "sed" command in curl was capable of.
- Worked with partner to go over questions and complete assignment.
- Although all those listed above contributed to my completing this work, all of its contents came from me alone.
- I used the website http://www.beardeddragonguide.com as the website I modified and took screen shots of.
References
- 13 Awesome Linux Grep Command Examples. (2017). Retrieved September 16, 2017, from https://tecadmin.net/grep-command-in-linux/
- Example Uses of Sed in Linux. (2017). Retrieved September 16, 2017, from https://www.lifewire.com/example-uses-of-sed-2201058
- LMU BioDB 2017. (2017). Week 3. Retrieved September 14, 2017, from https://xmlpipedb.cs.lmu.edu/biodb/fall2017/index.php/Week_3
Simonwro120 (talk) 03:35, 18 September 2017 (PDT)