Difference between revisions of "Jcowan4 Journal Week 8"
(→Methods/Results: added layout) |
(added the second step) |
||
| Line 38: | Line 38: | ||
#* Excel will now only display the rows that correspond to data meeting that filtering criterion. A number will appear in the lower left hand corner of the window giving you the number of rows that meet that criterion. We will check our results with each other to make sure that the computations were performed correctly. | #* Excel will now only display the rows that correspond to data meeting that filtering criterion. A number will appear in the lower left hand corner of the window giving you the number of rows that meet that criterion. We will check our results with each other to make sure that the computations were performed correctly. | ||
#* Be sure to undo any filters that you have applied before making any additional calculations. | #* Be sure to undo any filters that you have applied before making any additional calculations. | ||
| + | |||
| + | ===Calculate the Bonferroni and p value Correction=== | ||
| + | |||
| + | ''Note: Be sure to undo any filters that you have applied before continuing with the next steps.'' | ||
| + | # Now we will perform adjustments to the p value to correct for the [https://xkcd.com/882/ multiple testing problem]. Label the next two columns to the right with the same label, wt_Bonferroni_p-value. | ||
| + | # Type the equation <code>=<wt_p-value>*6189</code>, Upon completion of this single computation, use the Step (10) trick to copy the formula throughout the column. | ||
| + | # Replace any corrected p value that is greater than 1 by the number 1 by typing the following formula into the first cell below the second wt_Bonferroni_p-value header: <code>=IF(wt_Bonferroni_p-value>1,1,wt_Bonferroni_p-value)</code>, where "wt_Bonferroni_p-value" refers to the cell in which the first Bonferroni p value computation was made. Use the Step (10) trick to copy the formula throughout the column. | ||
| + | |||
==Data & Files== | ==Data & Files== | ||
Revision as of 13:03, 24 October 2019
Contents
Purpose
The purpose was to be able to conduct the "analyze" step of the data life cycle for a DNA microarray dataset (used to make sure data values are accurate), to develop an intuition about what different p-value cut-offs mean (understand the ratios and what fits a category) and to keep a detailed electronic laboratory notebook to facilitate reproducible research(to instruct any person to get the same exact results we got.
Methods/Results
Part 1
- We created a new worksheet, named it "wt_ANOVA"
- We copied the first three columns containing the "MasterIndex", "ID", and "Standard Name" from the "Master_Sheet" worksheet for our strain and pasted it into the worksheet. Then we copied the columns containing the data for our strain and pasted it into the worksheet.
- At the top of the first column to the right of our data, we created five column headers of the form wt_AvgLogFC_(15,30,60,90,120)
- In the cell below the wt_AvgLogFC_t15 header, we type "=AVERAGE"
- Then we highlighted all the data in row 2 associated with t15, then we pressed the enter (This cell now contains the average of the log fold change data from the first gene at t=15 minutes.)
- Click on this cell and position your cursor at the bottom right corner. You should see your cursor change to a thin black plus sign. When it does, double click, and the formula will be copied to the entire column of 6188 others genes.
- We repeated steps (4) through (8) with the t30, t60, t90, and the t120 data.
- Now in the first empty column to the right of the wt_AvgLogFC_t120 calculation, we created the column header wt_ss_HO.
- In the first cell below this header, we typed "=SUMSQ"
- Then highlighted all the LogFC data in row 2 (but not the AvgLogFC), press the closing paren key (shift 0),and press the "enter" key.
- In the next empty column to the right of wt_ss_HO, create the column headers wt_ss_(TIME) as in (3).
- Make a note of how many data points you have at each time point for your strain. For most of the strains, it will be 4, but for dHAP4 t90 or t120, it will be "3", and for the wild type it will be "4" or "5". Count carefully. Also, make a note of the total number of data points. Again, for most strains, this will be 20, but for example, dHAP4, this number will be 18, and for wt it should be 23 (double-check).
- In the first cell below the header wt_ss_t15, type
=SUMSQ(<range of cells for logFC_t15>)-COUNTA(<range of cells for logFC_t15>)*<AvgLogFC_t15>^2and hit enter.- The
COUNTAfunction counts the number of cells in the specified range that have data in them (i.e., does not count cells with missing values). - The phrase <range of cells for logFC_t15> should be replaced by the data range associated with t15.
- The phrase <AvgLogFC_t15> should be replaced by the cell number in which you computed the AvgLogFC for t15, and the "^2" squares that value.
- Upon completion of this single computation, use the Step (7) trick to copy the formula throughout the column.
- The
- Repeat this computation for the t30 through t120 data points. Again, be sure to get the data for each time point, type the right number of data points, and get the average from the appropriate cell for each time point, and copy the formula to the whole column for each computation.
- In the first column to the right of wt_ss_t120, create the column header wt_SS_full.
- In the first row below this header, type
=sum(<range of cells containing "ss" for each timepoint>)and hit enter. - In the next two columns to the right, create the headers wt_Fstat and wt_p-value.
- Recall the number of data points from (13): call that total n.
- In the first cell of the wt_Fstat column, type
=((n-5)/5)*(<wt_ss_HO>-<wt_SS_full>)/<wt_SS_full>and hit enter.- Don't actually type the n but instead use the number from (13). Also note that "5" is the number of timepoints.
- Replace the phrase wt_ss_HO with the cell designation.
- Replace the phrase <wt_SS_full> with the cell designation.
- Copy to the whole column.
- In the first cell below the wt_p-value header, type
=FDIST(<wt_Fstat>,5,n-5)replacing the phrase <wt_Fstat> with the cell designation and the "n" as in (13) with the number of data points total. . Copy to the whole column. - Before we move on to the next step, we will perform a quick sanity check to see if we did all of these computations correctly.
- Click on cell A1 and click on the Data tab. Select the Filter icon (looks like a funnel). Little drop-down arrows should appear at the top of each column. This will enable us to filter the data according to criteria we set.
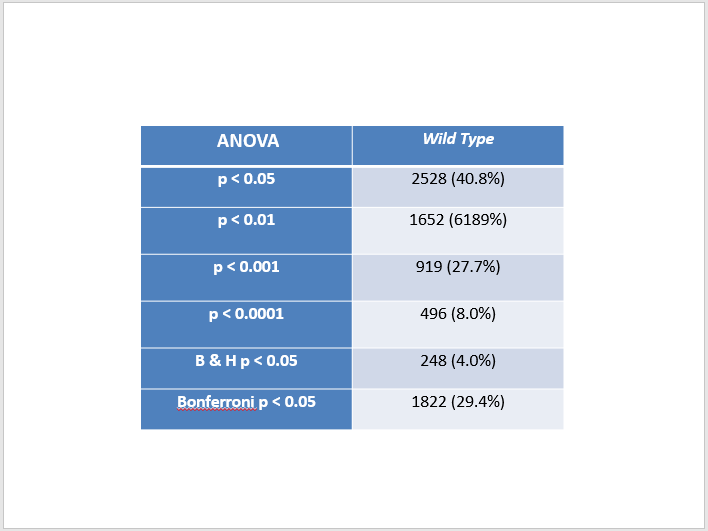
- Click on the drop-down arrow on your wt_p-value column. Select "Number Filters". In the window that appears, set a criterion that will filter your data so that the p value has to be less than 0.05.
- Excel will now only display the rows that correspond to data meeting that filtering criterion. A number will appear in the lower left hand corner of the window giving you the number of rows that meet that criterion. We will check our results with each other to make sure that the computations were performed correctly.
- Be sure to undo any filters that you have applied before making any additional calculations.
Calculate the Bonferroni and p value Correction
Note: Be sure to undo any filters that you have applied before continuing with the next steps.
- Now we will perform adjustments to the p value to correct for the multiple testing problem. Label the next two columns to the right with the same label, wt_Bonferroni_p-value.
- Type the equation
=<wt_p-value>*6189, Upon completion of this single computation, use the Step (10) trick to copy the formula throughout the column. - Replace any corrected p value that is greater than 1 by the number 1 by typing the following formula into the first cell below the second wt_Bonferroni_p-value header:
=IF(wt_Bonferroni_p-value>1,1,wt_Bonferroni_p-value), where "wt_Bonferroni_p-value" refers to the cell in which the first Bonferroni p value computation was made. Use the Step (10) trick to copy the formula throughout the column.
Data & Files
Conclusion
References
- Week 8. Retrieved October 15, 2019, from https://xmlpipedb.cs.lmu.edu/biodb/fall2019/index.php/Week_8
Acknowledgments
1. I worked with Kaitlyn, Christina, and Marcus for this assignment.
2. Dr. Dahlquist for the continued support in assisting and answering our questions during the making of this assignment.
3."Except for what is noted above, this individual journal entry was completed by me and not copied from another source."