Difference between revisions of "MSymond1 Week 5"
Jump to navigation
Jump to search
(→Questions: changed pixels and questions 1 and 2) |
(added template) |
||
| (3 intermediate revisions by the same user not shown) | |||
| Line 14: | Line 14: | ||
==Acknowledgements== | ==Acknowledgements== | ||
| − | I texted my homework partner, Charlotte, | + | I texted my homework partner, Charlotte, about a few of the questions for the assignment to help us understand the questions and the readings better. Except for what is noted above, this individual journal entry was completed by me and not copied from another source. [[User:Msymond1|Msymond1]] ([[User talk:Msymond1|talk]]) 21:58, 14 February 2024 (PST) |
==References== | ==References== | ||
| + | *Alberts et al. (2002) Molecular Biology of the Cell, Ch. 8: Microarrays | ||
| + | *Brown, P.O. & Botstein, D. (1999) Exploring the new world of the genome with DNA microarrays Nature Genetics 21: 33-37. https://doi.org/10.1038/4462 | ||
| + | *Campbell, A.M. and Heyer, L.J. (2003), “Chapter 4: Basic Research with DNA Microarrays”, in Discovering Genomics, Proteomics, and Bioinformatics, Cold Spring Harbor Laboratory Press, pp. 107-124. (Available on Brightspace) | ||
| + | *DeRisi, J.L., Iyer, V.R., and Brown, P.O. (1997) Exploring the Metabolic and Genetic Control of Gene Expression on a Genomic Scale. Science 278: 680-686. DOI: https://doi.org/10.1126/science.278.5338.680 | ||
| + | *LMU BioDB 2024. (2024). Week 5. Retrieved February 14, 2024, from https://xmlpipedb.cs.lmu.edu/biodb/spring2024/index.php/Week_5 | ||
| + | |||
| + | ==Template== | ||
| + | {{Template:MSymond1}} | ||
Latest revision as of 22:03, 14 February 2024
Contents
Purpose
The purpose of this assignment is to learn about how DNA microarrays work to sequence genomes. This assignment will include working through some examples of microarrays as well as uncovering times in which the ethics of data collection were violated.
Questions
- See Figure 1
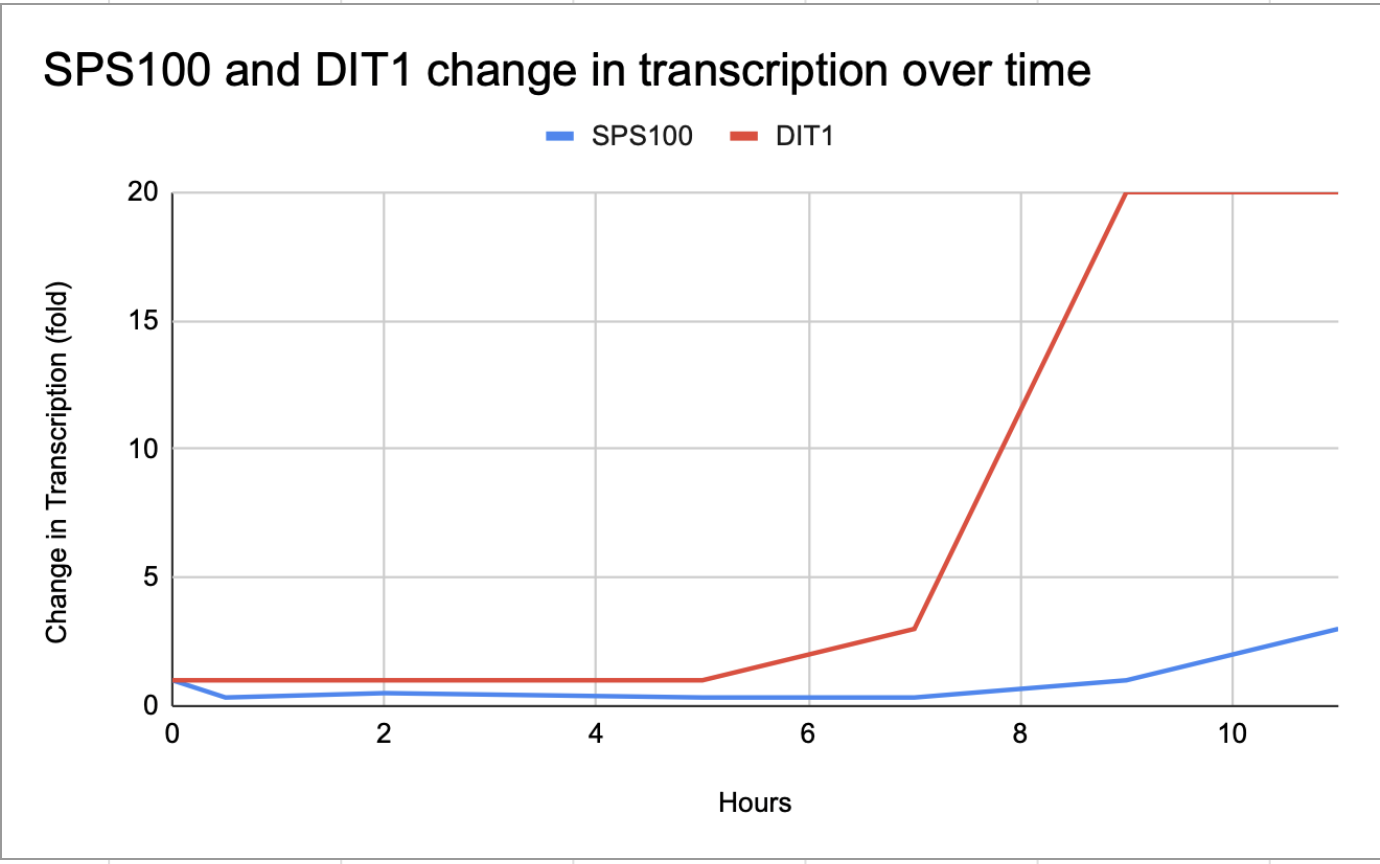
- See Figure 2
- Genes X and Y transcribed somewhat similarly. They both started at 1 and then increased, and then decreased down to about 1, and then close to 0
- The spots would be yellow at the first time point because they have not had any repression or induction yet. The technical reason for the spots showing up as yellow is the fact that they have not had any change since the starting point in their induction or transgression. The yellow color comes from the fact that it is what is in between the colors red and green, which are the colors that represent induction and transgression respectively. The biological reason that most genes ended up as yellow is because most genes do not get transgressed or induced or code for proteins.
- The TEF4 gene was repressed in the process of the experiment. It would make sense that the cell would turn green and become repressed in the absence of glucose because without its source of food, it has less energy to carry out functions with the DNA.
- The TCA cycle genes may have been induced when the glucose levels were decreasing because they sensed that the glucose levels were decreasing and thought that their energy sources would soon deplete. Therefore they rapidly induced through the course of the experiment.
- The genome could moderate the pathway to only allow the genes to grow when the conditions are right for them to induce or repress so they will all be in their own unique optimal conditions.
- The genes that would normally be repressed by the decrease in glucose would likely remain induced in this scenario without the repressor present. For this reason, the decrease in glucose would be associated with more induction and the spots would turn red.
- During an experiment with an over expression of the transcription factor YAP1p, the spots would turn into an intense red, for the YAP1p is what is responsible for the repression of glucose repressed genes. Meaning that this experiment would lead the spots to be induced. Leading to a red color.
- To test the data from the experiments, the modified samples with a deleted TUP1 or over expressed YAP1p could be compared to a control group of samples that have undergone no deletion or over expression.
Acknowledgements
I texted my homework partner, Charlotte, about a few of the questions for the assignment to help us understand the questions and the readings better. Except for what is noted above, this individual journal entry was completed by me and not copied from another source. Msymond1 (talk) 21:58, 14 February 2024 (PST)
References
- Alberts et al. (2002) Molecular Biology of the Cell, Ch. 8: Microarrays
- Brown, P.O. & Botstein, D. (1999) Exploring the new world of the genome with DNA microarrays Nature Genetics 21: 33-37. https://doi.org/10.1038/4462
- Campbell, A.M. and Heyer, L.J. (2003), “Chapter 4: Basic Research with DNA Microarrays”, in Discovering Genomics, Proteomics, and Bioinformatics, Cold Spring Harbor Laboratory Press, pp. 107-124. (Available on Brightspace)
- DeRisi, J.L., Iyer, V.R., and Brown, P.O. (1997) Exploring the Metabolic and Genetic Control of Gene Expression on a Genomic Scale. Science 278: 680-686. DOI: https://doi.org/10.1126/science.278.5338.680
- LMU BioDB 2024. (2024). Week 5. Retrieved February 14, 2024, from https://xmlpipedb.cs.lmu.edu/biodb/spring2024/index.php/Week_5
Template
User Page
Assignment Pages
Individual Journal Pages
- MSymond1 Week 1
- MSymond1 Week 2
- MSymond1 KMill104 Week 3
- NeMO_Week4
- MSymond1 Week 5
- MSymond1 Week 6
- MSymond1 Week 8
- MSymond1 Week 9
- MSymond1 Week 10
- MSymond1 Week 12
- MSymond1 Week 13
- MSymond1 Week 15