Difference between revisions of "Kmill104 Week 2"
(→References: adding first reference) |
(→Acknowledgements: adding final acknowledgements statement) |
||
| (6 intermediate revisions by the same user not shown) | |||
| Line 29: | Line 29: | ||
The aromatic amino acids must be in a specific position of the amino acid sequence to ensure its interactions with the amino acids around it will still result in color expression. In the examples above, the aromatic amino acid can only be found in the 10th position, the 11th position, or in both. If there is a disruption at other positions of the sequence, this may interfere with how the peptide sequence interacts and color could not be expressed even though an aromatic amino acid is present. | The aromatic amino acids must be in a specific position of the amino acid sequence to ensure its interactions with the amino acids around it will still result in color expression. In the examples above, the aromatic amino acid can only be found in the 10th position, the 11th position, or in both. If there is a disruption at other positions of the sequence, this may interfere with how the peptide sequence interacts and color could not be expressed even though an aromatic amino acid is present. | ||
| − | e. How do the colors combine to produce an overall color? How does this explain the genotype-phenotype rules you found in part (I)? | + | e. How do the colors combine to produce an overall color? How does this explain the genotype-phenotype rules you found in part (I)? Colors that only have one kind of allele and are then self-crossed will only result in that color. This is seen by self-crossing Green-1, which only has the one green allele and only results in the green color being expressed. Conversely, colors that have two alleles are then self-crossed may result in multiple colors. This is seen by self-crossing Green-2, which has two alleles, and results in green, yellow, and blue flowers. True breeding colors crossed with another true breeding color can also produce new colors, like green and white being crossed to created yellow and blue. So, overall color expression is dependent on the difference in alleles of the crossed organisms. |
| + | A specific genotype will only produce one phenotype, but the same phenotype may be observed across genotypes. | ||
| + | |||
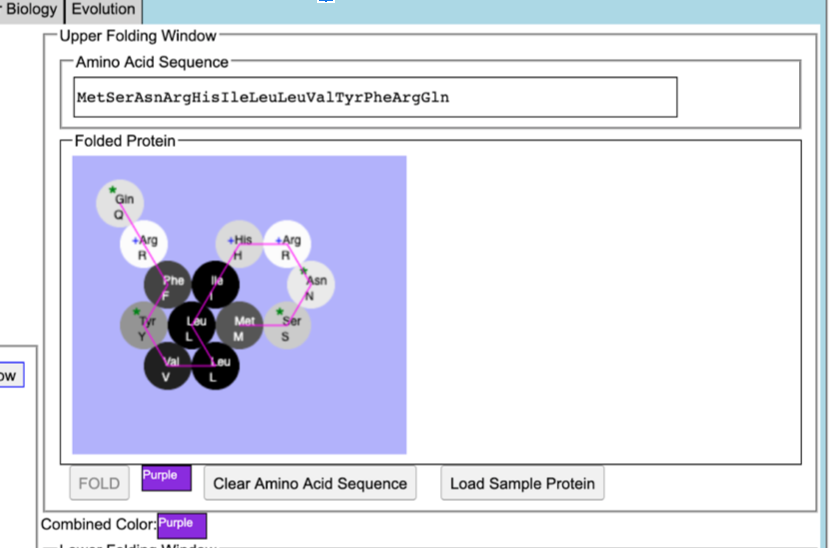
| + | f. Show your purple protein and explain why it is purple. The protein is purple because it contains two aromatic amino acids in its 10th and 11th positions, Tyr and Phe. The aromatic rings have a greater level of absorbance due to the ability of the ring electrons to move around their bonds, so the color purple is able to be expressed. | ||
| − | |||
[[image:Purpleprotein.png]] | [[image:Purpleprotein.png]] | ||
=== Scientific Conclusion === | === Scientific Conclusion === | ||
| + | This assignment showed that color expression is dependent on how a protein is structured, and specifically by its amino acid sequence. In order to absorb and emit UV wavelengths as a flower color, the sequence must contain a high number of alternating double bonds so that UV absorbance is high. Additionally, it is not always possible to determine the phenotypic result of crossing one or two organisms, as the many alleles involved in this process may produce one or multiple colored flowers. This shows us that phenotype cannot always help us determine a scientific result, and it is incredibly important to examine genetic material when we are looking for answers to scientific questions. | ||
=== Acknowledgements === | === Acknowledgements === | ||
| − | I worked with my homework partner Charlotte Kaplan [[User:Ckapla12]] to complete this assignment. We | + | I worked with my homework partner Charlotte Kaplan [[User:Ckapla12]] to complete this assignment. We texted twice to ask questions about formatting and about information we went over in class. |
| + | |||
| + | Except for what is noted above, this individual journal entry was completed by me and not copied from another source. | ||
=== References === | === References === | ||
LMU BioDb 2024. (2024). Week 2. Retrieved January 24, 2024, from | LMU BioDb 2024. (2024). Week 2. Retrieved January 24, 2024, from | ||
https://xmlpipedb.cs.lmu.edu/biodb/spring2024/index.php/Week_2 | https://xmlpipedb.cs.lmu.edu/biodb/spring2024/index.php/Week_2 | ||
| + | |||
| + | Aipotu. (2024). Retrieved January 24, 2024, from https://aipotu.umb.edu/ | ||
[[Template:Kmill104]] | [[Template:Kmill104]] | ||
{{Kmill104}} | {{Kmill104}} | ||
Latest revision as of 18:04, 24 January 2024
Contents
Purpose
The purpose of this assignment was to examine how specific color expression is determined in flowers. This was done by looking through the multiple scientific lenses of molecular biology, biochemistry, and genetics. Specifically, this assignment is meant to show that a precise amino acid sequence is necessary for a desired phenotypic result.
Method/Results
Protocol
Our first step was to start Aipotu and select the "Biochemistry" tab. We then practiced writing an amino acid sequence by using the individual letters, and then clicking "Fold" to see how our sequence folded. We examined each flower that was already present in the greenhouse and documented their sequence in our question responses below. We discovered additional colors by selecting the "Genetics" tab and either self-crossing one organism or crossing two organisms. Through this process, we were able to find blue, yellow, purple, orange, and black in addition to the preset green, red, and white flowers. We then observed the differences between their amino acid sequences to determine what amino acids a sequence must have to express any color besides white. By testing the different combinations of these amino acids we were able to find the sequence for a purple flower.
Question Responses
a. Which proteins are found in each of the four starting organisms?
- Green-1: MetSerAsnArgHisIleLeuLeuValTyrTrpArgGln
- Green-2: Combination of two alleles
- Blue allele has MetSerAsnArgHisIleLeuLeuValTyrCysArgGln
- Yellow allele has Trp replacing Tyr from the Blue allele
- Red: MetSerAsnArgHisIleLeuLeuValPheCysArgGln
- White: MetSerAsnArgHisIleLeuLeuValValCysArgGln
b. List the allele, color, and amino acid sequence
- allele: Green, color: Green, sequence: MetSerAsnArgHisIleLeuLeuVal(TyrTrp)ArgGln
- allele: Blue, color: Blue, sequence: MetSerAsnArgHisIleLeuLeuVal(TyrCys)ArgGln
- allele: Yellow, color: Yellow, sequence: MetSerAsnArgHisIleLeuLeuVal(TrpCys)ArgGln
- allele: Red, color: Red, sequence: MetSerAsnArgHisIleLeuLeuVal(PheCys)ArgGln
- allele: White, color: White, sequence: MetSerAsnArgHisIleLeuLeuVal(ValCys)ArgGln
- allele: Purple, color: Purple, sequence: MetSerAsnArgHisIleLeuLeuVal(TyrPhe)ArgGln
any difference in amino acid sequence is in parantheses
c. What features of a protein make it colored? A protein must contain at least one aromatic amino acid which increases the amount of alternating double bonds in the protein structure, allowing for a greater absorbance of UV light. A higher absorbance of UV light means the amino acids will then fluoresce and express some color.
d. What features of the amino acid sequence make a protein a particular color? The aromatic amino acids must be in a specific position of the amino acid sequence to ensure its interactions with the amino acids around it will still result in color expression. In the examples above, the aromatic amino acid can only be found in the 10th position, the 11th position, or in both. If there is a disruption at other positions of the sequence, this may interfere with how the peptide sequence interacts and color could not be expressed even though an aromatic amino acid is present.
e. How do the colors combine to produce an overall color? How does this explain the genotype-phenotype rules you found in part (I)? Colors that only have one kind of allele and are then self-crossed will only result in that color. This is seen by self-crossing Green-1, which only has the one green allele and only results in the green color being expressed. Conversely, colors that have two alleles are then self-crossed may result in multiple colors. This is seen by self-crossing Green-2, which has two alleles, and results in green, yellow, and blue flowers. True breeding colors crossed with another true breeding color can also produce new colors, like green and white being crossed to created yellow and blue. So, overall color expression is dependent on the difference in alleles of the crossed organisms. A specific genotype will only produce one phenotype, but the same phenotype may be observed across genotypes.
f. Show your purple protein and explain why it is purple. The protein is purple because it contains two aromatic amino acids in its 10th and 11th positions, Tyr and Phe. The aromatic rings have a greater level of absorbance due to the ability of the ring electrons to move around their bonds, so the color purple is able to be expressed.
Scientific Conclusion
This assignment showed that color expression is dependent on how a protein is structured, and specifically by its amino acid sequence. In order to absorb and emit UV wavelengths as a flower color, the sequence must contain a high number of alternating double bonds so that UV absorbance is high. Additionally, it is not always possible to determine the phenotypic result of crossing one or two organisms, as the many alleles involved in this process may produce one or multiple colored flowers. This shows us that phenotype cannot always help us determine a scientific result, and it is incredibly important to examine genetic material when we are looking for answers to scientific questions.
Acknowledgements
I worked with my homework partner Charlotte Kaplan User:Ckapla12 to complete this assignment. We texted twice to ask questions about formatting and about information we went over in class.
Except for what is noted above, this individual journal entry was completed by me and not copied from another source.
References
LMU BioDb 2024. (2024). Week 2. Retrieved January 24, 2024, from https://xmlpipedb.cs.lmu.edu/biodb/spring2024/index.php/Week_2
Aipotu. (2024). Retrieved January 24, 2024, from https://aipotu.umb.edu/
User Page
Assignment Pages
- Week 1
- Week 2
- Week 3
- Week 4
- Week 5
- Week 6
- Week 8
- Week 9
- Week 10
- Week 11
- Week 12
- Week 13
- Week 14
- Week 15
Individual Journal Entry Pages
- Kmill104 Week 1
- Kmill104 Week 2
- MSymond1 KMill104 Week 3
- Monarch Initiative Week 4
- Kmill104 Week 5
- Kmill104 Week 6
- Kmill104 Week 8
- Kmill104 Week 9
- Kmill104 Week 10
- Kmill104 Week 11
- Kmill104 Week 12
- Data Analysts Week 13
- Data Analysts Week 14
- Data Analysts Week 15