Difference between revisions of "MSymond1 Week 2"
(finished acknowledgements) |
(added references) |
||
| Line 27: | Line 27: | ||
[[https://en.wikipedia.org/wiki/Help:Wikitext#:~:text=The%20markup%20language%20called%20wikitext,edit%2C%20see%20Help%3AEditing.| Click here]] | [[https://en.wikipedia.org/wiki/Help:Wikitext#:~:text=The%20markup%20language%20called%20wikitext,edit%2C%20see%20Help%3AEditing.| Click here]] | ||
Except for what is noted above, this individual journal entry was completed by me and not copied from another source. [[User:Msymond1|Msymond1]] ([[User talk:Msymond1|talk]]) 22:34, 24 January 2024 (PST) | Except for what is noted above, this individual journal entry was completed by me and not copied from another source. [[User:Msymond1|Msymond1]] ([[User talk:Msymond1|talk]]) 22:34, 24 January 2024 (PST) | ||
| + | =======References====== | ||
| + | LMU BioDb 2024. (2024). Week 2. Retrieved January 24, 2024, from | ||
| + | https://xmlpipedb.cs.lmu.edu/biodb/spring2024/index.php/Week_2 | ||
| + | |||
| + | Aipotu. (2024). Retrieved January 24, 2024, from https://aipotu.umb.edu/ | ||
*{{Template:MSymond1}} | *{{Template:MSymond1}} | ||
Revision as of 22:36, 24 January 2024
Contents
Purpose
The purpose of this lab is to study what has been found in the genetics and biochemistry sections of the lab, but diving deeper into the molecular biology of the different colors, and to determine what differences in the DNA sequences lead to the differences in phenotypes seen when the flowers are crossed either with themselves or with others.
Methods and Results
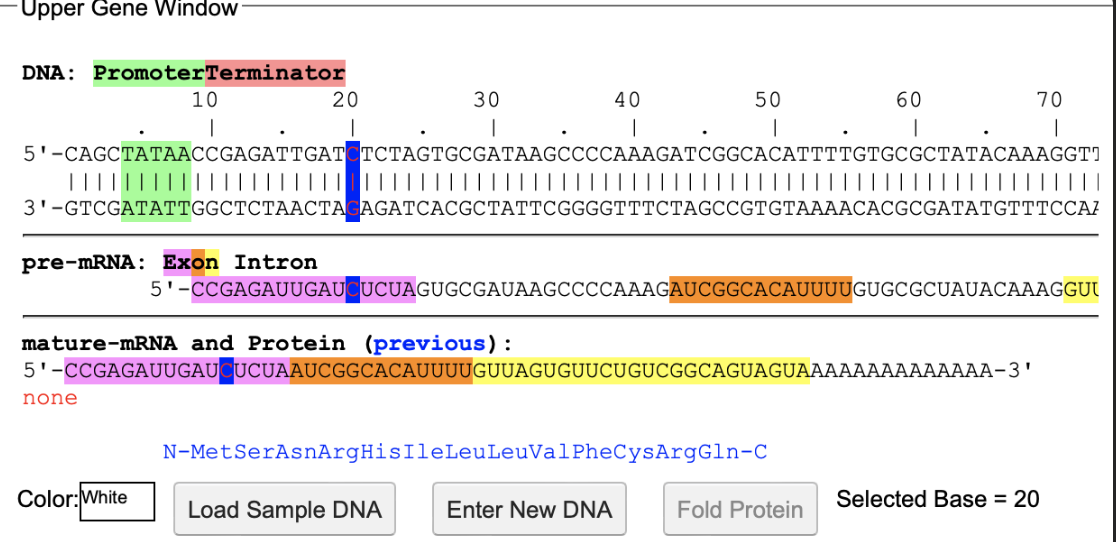
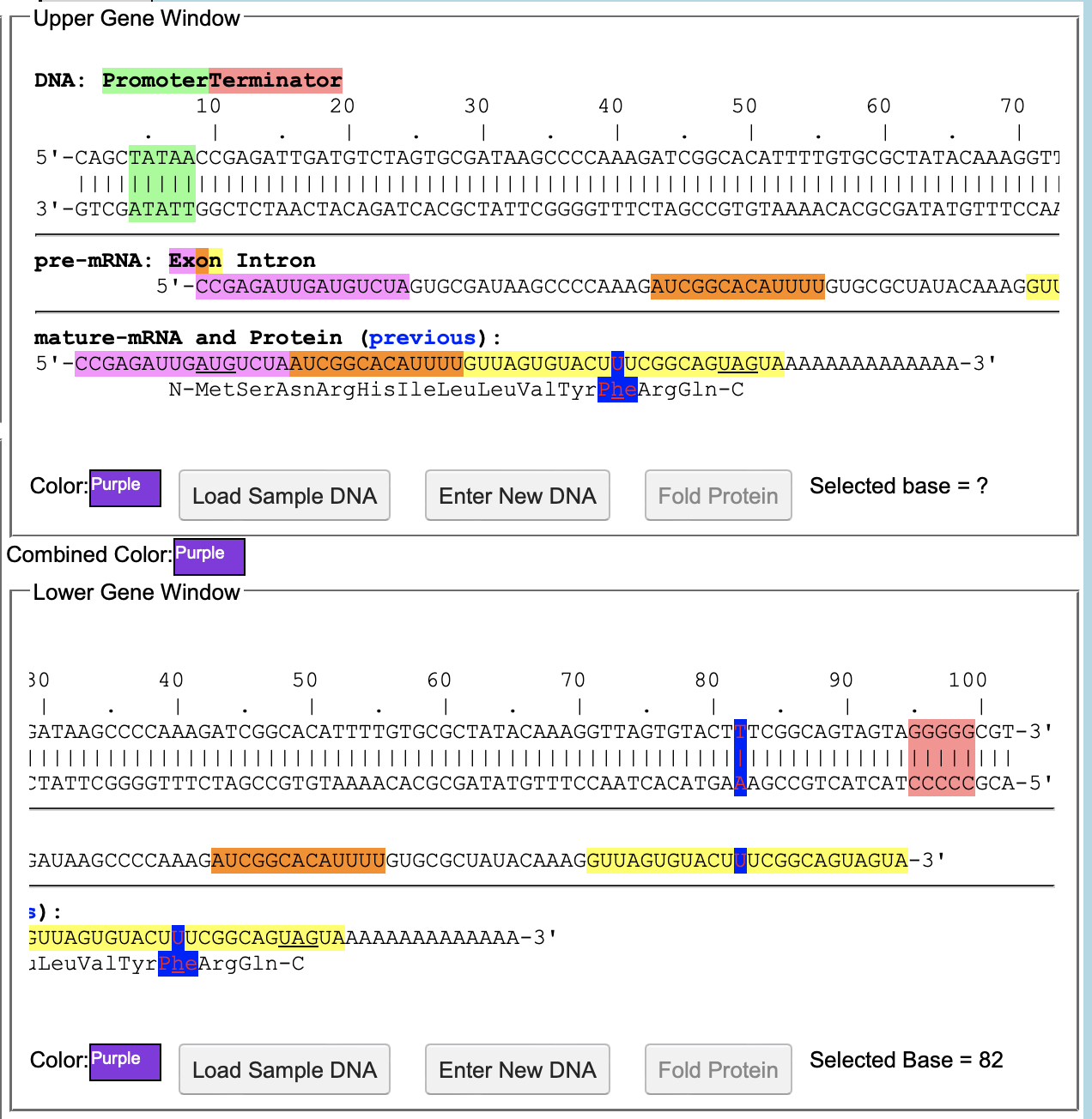
The software Aipotu was used for this lab, using the molecular biology tab. The completion of the molecular biology section of this lab did require assistance from the group that completed the genetics portion and the biochemistry portion. They helped provide a list of all different alleles, as well as which alleles are true breeding. The molecular biology tab of this software displays the entire DNA and protein sequence of the alleles and what color pigment they make. The first DNA sequence edited in this experiment was the sequence from the red flower. The base that was edited was from the red sequence, and the base was base #20. It was changed from a G to a C. This led the protein sequence to not be folded, because it did not allow for methionine to be coded, therefore, no pigmented protein could be created, making it a white flower. This led to the discovery that there are multiple ways to create a white flower, because any DNA sequence that does not create a functional pigmented protein will create a white flower (figure 1). Using the knowledge of the different alleles, provided by the genetics group, as well as the biochemistry of a purple allele from the biochemistry group, a purple allele for flowers was created using the molecular biology of the organisms to create a true breeding purple organism (figure 2). The three aromatic amino acids that are responsible for color are phenylalanine, tyrosine, and tryptophan. The red sequence from a purple heterozygote had phenylalanine, and the blue sequence had tyrosine. So an amino acid sequence was created that had both of those amino acids. This was done by changing base 82 of a blue sequence to a U, giving it a phenylalanine.
Specific Tasks
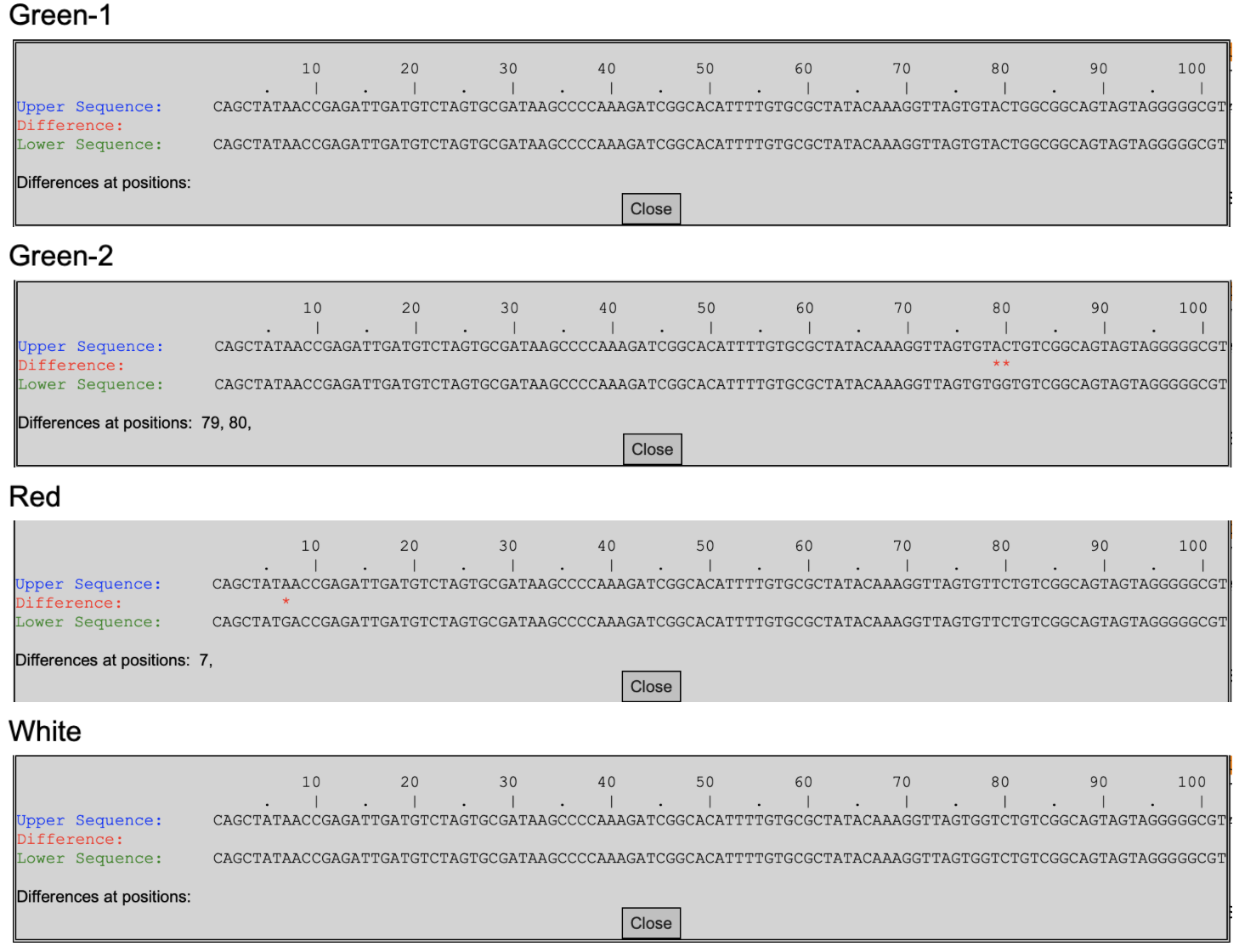
A) There are no differences in the Green DNA sequences in the Green-1 flower. The differences between blue and yellow sequences in Green-2 were at positions 79, and 80. The differences between the red and white sequences in the red flower are at position 7. There were no differences between the white sequences in the white flower. Between every available allele of the colored (non-white) flowers, all of the differences in the DNA sequences fell in bases 79 and 80
- See figure 3
B) Not all white DNA sequences are the same. When a base was changed on the methionine codon, the protein sequence could not be folded, and the flower turned white. Meaning that whenever a protein sequence fails to be folded, it turns into a white flower, because there is no pigmentations
C) Green-1 was Green and Green. Green-2 was Blue and Yellow. Red was Red and White. White was White and White
- See figure 4
D) The Purple DNA sequence was created by changing base 82 to a U, adding a phenylalanine to the sequence.
- See figure 5
E) Certain changes made to the DNA sequences could explain the mutations seen in part one because certain changes to DNA sequences included deletions or insertions, which in some cases was the deletion of methionine, which meant that there was no pigmented protein at all, which led to a white flower. This could also explain some of the mutations in part one that led to a change in color, for they could have changed one of the bases at positions 79 or 80 or they could have changed one of the proteins to one of the aromatic proteins to alter the color of the flower.
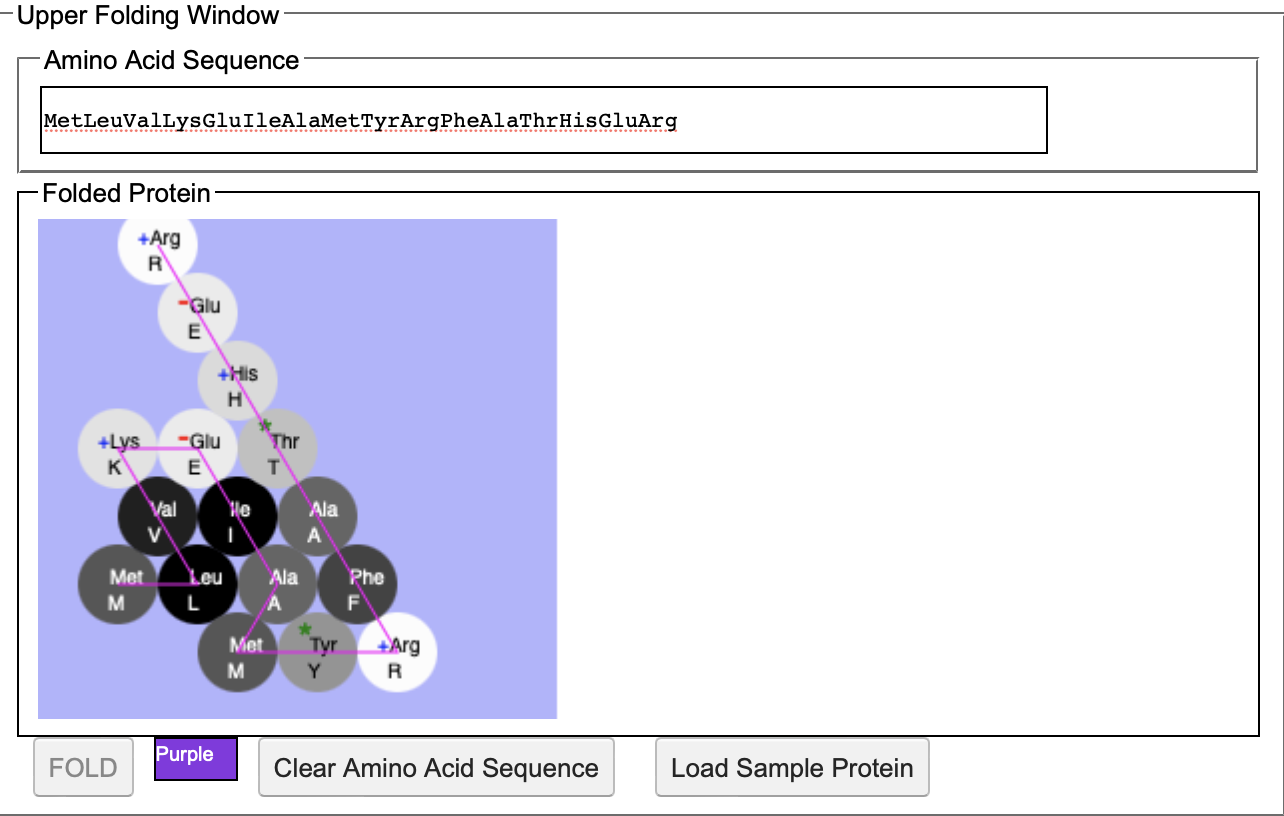
F) The protein creates a purple pigmentation.
- See figure 6
Scientific Conclusion
This lab showed significant implications for the molecular underpinnings for the pigmentation of the flowers found in parts I and II of the lab. After determining all possible alleles for the flowers, and which proteins are found to be different in the different alleles, this part of the lab was able to create an organism of a new allele for the color purple. After it was determined that the only difference between the red and blue DNA sequences (the sequences that made the purple heterozygote flower) was on base 79, and that there was one difference in proteins between the sequences, a sequence was created that had both of the proteins that set apart the DNA sequences.
Acknowledgements
I worked with my homework partner Natalija Stojanovic. Although she was not present in class this week on Tuesday, we exchanged texts and called for 15 minutes. I helped clarify a few things about how to use the software to complete the assignment. I also consulted the homework partners who were working on the biochemistry section of the lab. I also consulted my professor Dr. Dahlquist during class, as she helped me determine the three aromatic amino acids. Wikipedia page that shows how to use "wikitext". [Click here] Except for what is noted above, this individual journal entry was completed by me and not copied from another source. Msymond1 (talk) 22:34, 24 January 2024 (PST)
=References
LMU BioDb 2024. (2024). Week 2. Retrieved January 24, 2024, from https://xmlpipedb.cs.lmu.edu/biodb/spring2024/index.php/Week_2
Aipotu. (2024). Retrieved January 24, 2024, from https://aipotu.umb.edu/
- ====User Page====
- User:Msymond1
Assignment Pages
Individual Journal Pages
- MSymond1 Week 1
- MSymond1 Week 2
- MSymond1 KMill104 Week 3
- NeMO_Week4
- MSymond1 Week 5
- MSymond1 Week 6
- MSymond1 Week 8
- MSymond1 Week 9
- MSymond1 Week 10
- MSymond1 Week 12
- MSymond1 Week 13
- MSymond1 Week 15