Difference between revisions of "Hivanson and Nstojan1 Week 3"
Jump to navigation
Jump to search
(Added answer to question) |
(edited syntax) |
||
| Line 34: | Line 34: | ||
**The molecular function of IMD3 is that it enables IMP dehydrogenase activity as well as mRNA binding. It catalyzes the rate-limiting step in the de novo synthesis of GTP and its involved in GTP biosynthesis pathways. In addition, it catalyzes the conversion of inosine 5'-phosphate (IMP) to xanthosine 5'-phosphate (XMP). It is also known to play an important roll in cell growth. | **The molecular function of IMD3 is that it enables IMP dehydrogenase activity as well as mRNA binding. It catalyzes the rate-limiting step in the de novo synthesis of GTP and its involved in GTP biosynthesis pathways. In addition, it catalyzes the conversion of inosine 5'-phosphate (IMP) to xanthosine 5'-phosphate (XMP). It is also known to play an important roll in cell growth. | ||
*What was different about the information provided about your gene in each of the parent databases? | *What was different about the information provided about your gene in each of the parent databases? | ||
| − | **SDG and Uniprot both provide a variety of in-depth information about the gene. The contents of their information is fairly similar, in particular UniProt has more visual images and charts/graphs compared to SDG. e!Ensemble contains generic information that just covers the basics such as a name, description and type. | + | ***SDG and Uniprot both provide a variety of in-depth information about the gene. The contents of their information is fairly similar, in particular UniProt has more visual images and charts/graphs compared to SDG. e!Ensemble contains generic information that just covers the basics such as a name, description and type. |
**Were there differences in content, the information or data itself? | **Were there differences in content, the information or data itself? | ||
**Were there differences in presentation of the information? | **Were there differences in presentation of the information? | ||
Revision as of 19:31, 31 January 2024
IMD3
Summary of Function:
Other Information:
- What is the standard name, systematic name, and name description for your gene (from SGD)?
- Standard name: IMD3
- Systematic name: YLR432W
- Name description: IMP Dehydrogenase
- What is the gene ID (identifier) for your gene in all four databases (SGD, NCBI Gene, Ensembl, UniProt)?
- What is the DNA sequence of your gene?
- ATGGCCGCCGTTAGAGACTACAAGACTGCCTTGGAATTCGCCAAGAGCTTACCAAGACTAGATGGTTTGTCTGTCCAGGAGTTGATGGACTCCAAGACCAGAGGTGGGTTGACTTATAACGACTTTTTGGTTTTGCCAGGTCTGGTTGATTTCCCATCTTCTGAAGTTAGCCTACAAACTAAGTTGACAAGGAATATCACTTTGAACACCCCATTCGTTTCCTCTCCAATGGACACCGTGACAGAATCAGAAATGGCCATCTTCATGGCTTTGTTGGGTGGTATCGGTTTTATTCACCACAACTGTACCCCAGAGGACCAAGCTGACATGGTCAGAAGAGTCAAGAACTATGAAAATGGGTTTATTAACAACCCTATAGTGATTTCCCCAACTACTACTGTGGGTGAAGCTAAGAGTATGAAGGAAAGATTTGGATTTTCCGGTTTCCCCGTTACAGAAGATGGTAAAAGAAACGGAAAATTGATGGGTATCGTCACTTCTCGTGATATTCAGTTCGTTGAAGACAACTCTTTGCTTGTTCAAGATGTTATGACCAAAAACCCTGTCACCGGTGCACAAGGTATTACATTGTCTGAAGGTAATGAAATTTTAAAGAAGATTAAAAAGGGTAAGCTATTGATTGTTGACGACAATGGTAACTTAGTTTCTATGCTTTCCAGAACTGATTTAATGAAAAATCAGAACTATCCATTGGCTTCTAAATCTGCCACCACCAAGCAACTGTTATGTGGTGCTGCTATCGGTACTATCGATGCTGATAAGGAAAGGTTAAGACTATTAGTCGAAGCAGGTTTGGATGTTGTTATCTTAGATTCCTCTCAAGGTAACTCTATTTTCCAATTGAACATGATCAAATGGATTAAAGAAACTTTCCCAGATTTGGAAATCATTGCTGGTAACGTTGCCACCAGAGAACAAGCTGCTAACTTGATTGCTGCCGGTGCCGATGGTTTAAGAATTGGTATGGGTTCAGGCTCTATTTGTATCACTCAAGAAGTTATGGCCTGTGGTAGACCACAAGGTACAGCCGTCTACAACGTGTGTGAATTTGCTAACCAATTCGGTATTCCATGTATGGCTGATGGTGGTGTTCAAAACATTGGTCATATTACCAAAGCTTTGGCTCTTGGTTCTTCTACTGTTATGATGGGTGGTATGTTAGCTGGTACTACTGAATCTCCTGGTGAATATTTCTATCAAGATGGTAAAAGATTGAAGGCATATCGTGGTATGGGTTCCATTGACGCCATGCAAAAGACTGGTACTAAAGGTAATGCATCTACCTCCCGTTACTTTTCCGAATCAGACAGTGTTTTGGTCGCACAAGGTGTCTCCGGTGCTGTCGTTGACAAAGGTTCTATCAAGAAATTTATTCCATATTTGTACAACGGTTTACAACATTCTTGTCAAGACATTGGTTACAAGTCCCTAACTTTATTAAAGGAAAATGTCCAAAGCGGTAAAGTTAGATTTGAATTTAGAACCGCTTCTGCTCAACTAGAAGGTGGTGTTCATAACTTACATTCTTACGAAAAGCGTTTACATAACTAA
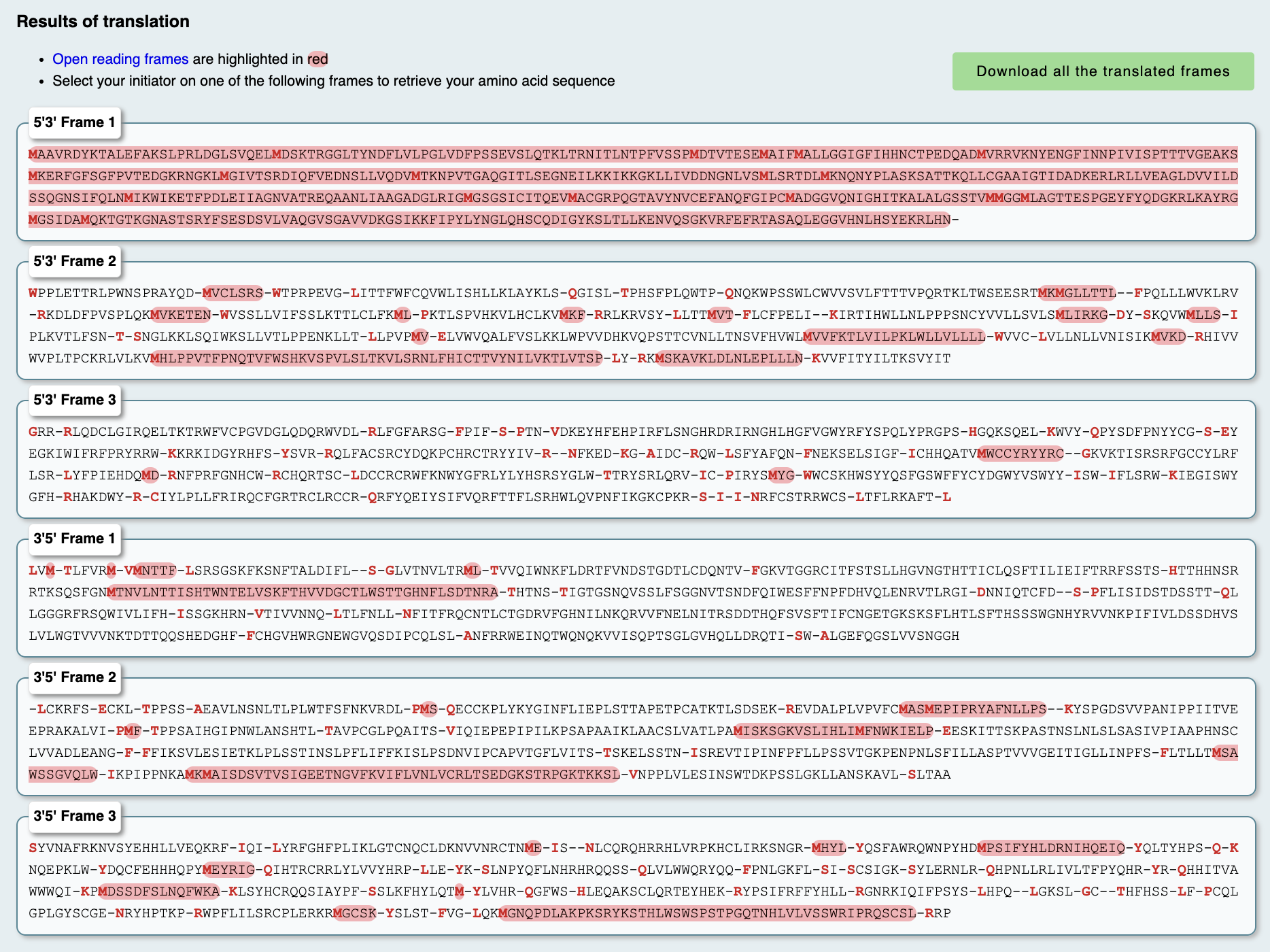
- What is the protein sequence corresponding to your gene?
Reading frame 1 encodes the IMD3 protein sequence:
MAAVRDYKTALEFAKSLPRLDGLSVQELMDSKTRGGLTYNDFLVLPGLVD FPSSEVSLQTKLTRNITLNTPFVSSPMDTVTESEMAIFMALLGGIGFIHH NCTPEDQADMVRRVKNYENGFINNPIVISPTTTVGEAKSMKERFGFSGFP VTEDGKRNGKLMGIVTSRDIQFVEDNSLLVQDVMTKNPVTGAQGITLSEG NEILKKIKKGKLLIVDDNGNLVSMLSRTDLMKNQNYPLASKSATTKQLLC GAAIGTIDADKERLRLLVEAGLDVVILDSSQGNSIFQLNMIKWIKETFPD LEIIAGNVATREQAANLIAAGADGLRIGMGSGSICITQEVMACGRPQGTA VYNVCEFANQFGIPCMADGGVQNIGHITKALALGSSTVMMGGMLAGTTES PGEYFYQDGKRLKAYRGMGSIDAMQKTGTKGNASTSRYFSESDSVLVAQG VSGAVVDKGSIKKFIPYLYNGLQHSCQDIGYKSLTLLKENVQSGKVRFEF RTASAQLEGGVHNLHSYEKRLHN-
- What is the function of your gene?
- The molecular function of IMD3 is that it enables IMP dehydrogenase activity as well as mRNA binding. It catalyzes the rate-limiting step in the de novo synthesis of GTP and its involved in GTP biosynthesis pathways. In addition, it catalyzes the conversion of inosine 5'-phosphate (IMP) to xanthosine 5'-phosphate (XMP). It is also known to play an important roll in cell growth.
- What was different about the information provided about your gene in each of the parent databases?
- SDG and Uniprot both provide a variety of in-depth information about the gene. The contents of their information is fairly similar, in particular UniProt has more visual images and charts/graphs compared to SDG. e!Ensemble contains generic information that just covers the basics such as a name, description and type.
- Were there differences in content, the information or data itself?
- Were there differences in presentation of the information?
- Why did you choose your particular gene? i.e., why is it interesting to you and your partner?
- Hailey’s research in Dr. Sarah Mitchell’s lab at LMU studies IMD3’s RNA binding function in yeast.
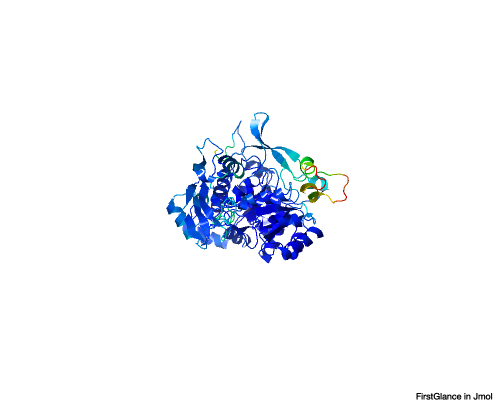
- Include an image related to your gene (be careful that you do not violate any copyright restrictions!)