Difference between revisions of "Hivanson and Nstojan1 Week 3"
Jump to navigation
Jump to search
(→IMD3: expasy is causing little issues idk why i didn't just preview last time.) |
(→Other Information:: answer why imd3) |
||
| Line 34: | Line 34: | ||
#*Were there differences in presentation of the information? | #*Were there differences in presentation of the information? | ||
#Why did you choose your particular gene? i.e., why is it interesting to you and your partner? | #Why did you choose your particular gene? i.e., why is it interesting to you and your partner? | ||
| + | #*Hailey’s research in Dr. Sarah Mitchell’s lab at LMU studies IMD3’s RNA binding function in yeast. | ||
#Include an image related to your gene (be careful that you do not violate any copyright restrictions!) | #Include an image related to your gene (be careful that you do not violate any copyright restrictions!) | ||
#*Please make the image something scientific (not like the random images seen on the SGD blog posts). | #*Please make the image something scientific (not like the random images seen on the SGD blog posts). | ||
#*If a 3D structure of the protein your gene encodes is available, you can choose to embed a rotating image of the structure on your page using the FirstGlance in Jmol software. This is optional, a different static image would be OK, too. | #*If a 3D structure of the protein your gene encodes is available, you can choose to embed a rotating image of the structure on your page using the FirstGlance in Jmol software. This is optional, a different static image would be OK, too. | ||
#*The NCBI Structure database and RSCB Protein Databank also display structures. | #*The NCBI Structure database and RSCB Protein Databank also display structures. | ||
Revision as of 18:41, 31 January 2024
IMD3
Summary of Function:
Other Information:
- What is the standard name, systematic name, and name description for your gene (from SGD)?
- Standard name: IMD3
- Systematic name: YLR432W
- Name description: IMP Dehydrogenase
- What is the gene ID (identifier) for your gene in all four databases (SGD, NCBI Gene, Ensembl, UniProt)?
- What is the DNA sequence of your gene?
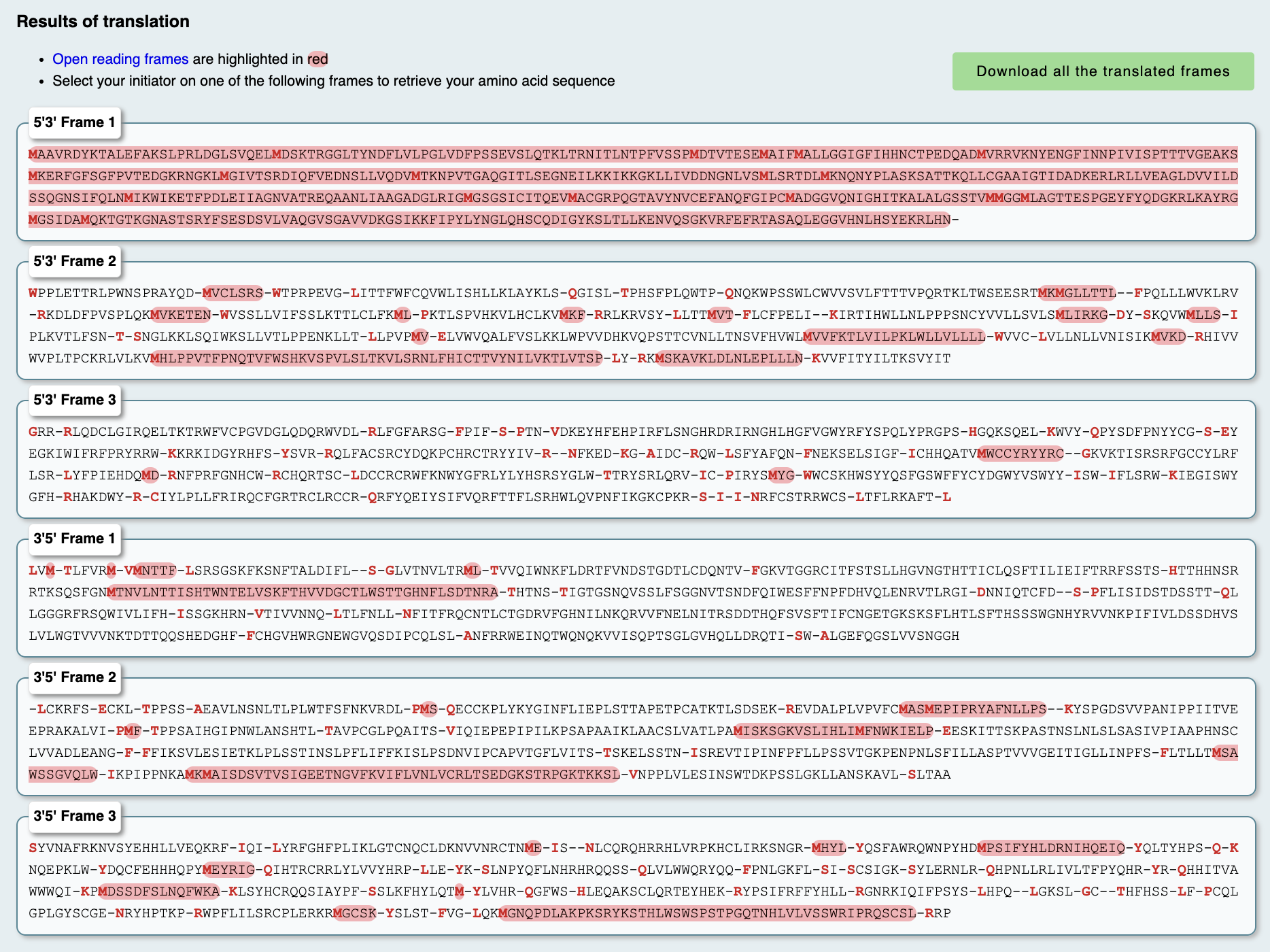
- What is the protein sequence corresponding to your gene?
Reading frame 1 encodes the IMD3 protein sequence:
MAAVRDYKTALEFAKSLPRLDGLSVQELMDSKTRGGLTYNDFLVLPGLVD FPSSEVSLQTKLTRNITLNTPFVSSPMDTVTESEMAIFMALLGGIGFIHH NCTPEDQADMVRRVKNYENGFINNPIVISPTTTVGEAKSMKERFGFSGFP VTEDGKRNGKLMGIVTSRDIQFVEDNSLLVQDVMTKNPVTGAQGITLSEG NEILKKIKKGKLLIVDDNGNLVSMLSRTDLMKNQNYPLASKSATTKQLLC GAAIGTIDADKERLRLLVEAGLDVVILDSSQGNSIFQLNMIKWIKETFPD LEIIAGNVATREQAANLIAAGADGLRIGMGSGSICITQEVMACGRPQGTA VYNVCEFANQFGIPCMADGGVQNIGHITKALALGSSTVMMGGMLAGTTES PGEYFYQDGKRLKAYRGMGSIDAMQKTGTKGNASTSRYFSESDSVLVAQG VSGAVVDKGSIKKFIPYLYNGLQHSCQDIGYKSLTLLKENVQSGKVRFEF RTASAQLEGGVHNLHSYEKRLHN-
- What is the function of your gene?
- What was different about the information provided about your gene in each of the parent databases?
- Were there differences in content, the information or data itself?
- Were there differences in presentation of the information?
- Why did you choose your particular gene? i.e., why is it interesting to you and your partner?
- Hailey’s research in Dr. Sarah Mitchell’s lab at LMU studies IMD3’s RNA binding function in yeast.
- Include an image related to your gene (be careful that you do not violate any copyright restrictions!)
- Please make the image something scientific (not like the random images seen on the SGD blog posts).
- If a 3D structure of the protein your gene encodes is available, you can choose to embed a rotating image of the structure on your page using the FirstGlance in Jmol software. This is optional, a different static image would be OK, too.
- The NCBI Structure database and RSCB Protein Databank also display structures.