Kmill104 Week 5
Jump to navigation
Jump to search
Contents
Purpose
The purpose of this assignment is to learn about DNA microarray experiments and how to read the data that results from this process. Analysis problems about microarray experiments are also given to better our understanding of the process and how we should derive meaning from the different colored spots. Finally, this assignment is also meant to show us a case where the ethics surrounding research were violated and what outcomes come from this and other cases like it.
Questions
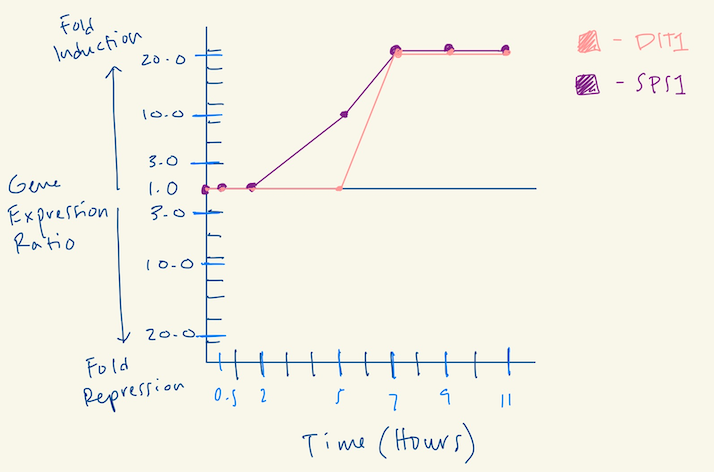
- (Question 5, p. 110) Choose two genes from Figure 4.6b (PDF of figures on Brightspace) and draw a graph to represent the change in transcription over time. You can either create your plot in Excel and put the image up on your wiki page or you can do it by hand and upload a picture or scan.
- (Question 6b, p. 110) Look at Figure 4.7, which depicts the loss of oxygen over time and the transcriptional response of three genes. These data are the ratios of transcription for genes X, Y, and Z during the depletion of oxygen. Using the color scale from Figure 4.6, determine the color for each ratio in Figure 4.7b. (Use the nomenclature "bright green", "medium green", "dim green", "black", "dim red", "medium red", or "bright red" for your answers.) For genes X, Y, and Z after 1 hour, they would all exhibit a black color. At the 3 hour mark, gene X would be a dim red, gene Y a medium red, and gene Z a dim red. At the 5 hour mark, gene x would be black, gene Y a dim green, and gene Z a dim red. At the 9 hour mark, gene X would be medium green, gene Y a bright green, and gene Z a dim red.
- (Question 7, p. 110) Were any of the genes in Figure 4.7b transcribed similarly? If so, which ones were transcribed similarly to which ones? Genes X, Y, and Z all had the same rate of transcription after 1 hour, but after this hour mark their responses varied. Each gene had an increased rate of transcription at the 3 hour mark, but the amount that they increased by was different. Gene Y had the largest increase in transcription, while Gene Z had the lowest. After the 3 hour mark, both Gene X and Gene Y saw a decrease in transcription. Gene X dropped from a 2.2 to a 1.0 ratio, while Gene Y dropped from 4.5 to 0.95. 1.0 and 0.95 are close in value so we see some similarities there, but at the 9 hour mark Gene Y again sees a dramatic decrease in transcription from 0.95 to 0.05, while Gene X drops from 1.0 to 0.15. Though the ranges of values for Gene X and Gene Y are different, both genes follow a similar trajectory for when their rate of transcription is highest and lowest. Conversely, Gene Z continuously increases in transcription, and between the 5 and 9 hour mark it seems to level out at a ratio of 2.0.
- (Question 9, p. 118) Why would most spots be yellow at the first time point? I.e., what is the technical reason that spots show up as yellow - where does the yellow color come from? And, what would be the biological reason that the experiment resulted in most spots being yellow? Yellow spots mean there is an equal ratio of red cDNA, which is the experimental cDNA, and green DNA, which is the control cDNA. If we get yellow spots from comparing the experimental and control cDNA, then there has been no change in the gene expression. At the first time point, the cells have not been exposed long enough to an environment lacking in glucose for there to be any change in gene expression. They likely have stores of glucose that they are able to use for transcription and the process occurs as normal. Transcription will be affected when they run out of any stored glucose and there is not enough glucose in their environment to replace it.
- (Question 10, p. 118) Go to the Saccharomyces Genome Database and search for the gene TEF4; you will see it is involved in translation. Look at the time point labeled OD 3.7 in Figure 4.12, and find the TEF4 spot. Over the course of this experiment, was TEF4 induced or repressed? Hypothesize why TEF4’s change in expression was part of the cell’s response to a reduction in available glucose (i.e., the only available food). TEF4 was repressed, as its spot color is slightly more green than yellow, indicating repression. If the cell does not have enough available glucose, then it cannot go through cellular respiration at the same rate in order to produce energy. Without energy, the cell cannot transcribe its DNA at the same rate into mRNA. And with a slower rate of mRNA production, there will also be a reduced rate of translation occurring. This is because transcription provides the mRNA strand that is read during translation and codes for a protein. If there is a reduced rate of translation, there is less of a need for TEF4 being involved in the process and its expression is repressed.
- (Question, 11, p. 120) Why would TCA cycle genes be induced if the glucose supply is running out? The TCA cycle genes producing proteins phosphoenolpyruvate carboxykinase and fructose 1,6-biphosphatase are induced, as they control the irreversible steps of glycolysis that need to be regulated in order to reverse the flow of carbon towards glucose. The proteins trehalose synthase and glycogen synthase are induced as well, as their function involves converting glucose into storage sugars that can later be used for energy.
- (Question 12, p. 120) What mechanism could the genome use to ensure genes for enzymes in a common pathway are induced or repressed simultaneously? Transcription factors for the gene are stimulated when there is not enough of the enzyme, and they go to induce transcription so that enough enzyme is formed. Conversely, if there is too much/enough of the enzyme in the cell, the transcription factor is not stimulated and does not induce transcription, so that energy isn't being wasted on forming something that is not needed. There could also be some factor that only induces, while another factor only represses, so that transcription can be equally induced and repressed.
- (Question 13, p. 121) Consider a microarray experiment where cells deleted for the repressor TUP1 were subjected to the same experiment of a timecourse of glucose depletion where cells at t0 (plenty of glucose available) are labeled green and cells at later timepoints (glucose depleted) are labeled red. What color would you expect the spots that represented glucose-repressed genes to be in the later time points of this experiment? You would expect the spots to be red, because when the repressor is deleted in cells that are in the glucose depleted environment, gene expression is induced and we see red spots. If there is no repressor to be activated by glucose, and there is no glucose present, then there will always be transcription occurring.
- (Question 14, p. 121) Consider a microarray experiment where cells that overexpress the transcription factor Yap1p were subjected to the same experiment of a timecourse of glucose depletion where cells at t0 (plenty of glucose available) are labeled green and cells at later timepoints (glucose depleted) are labeled red. What color would you expect the spots that represented Yap1p target genes to be in the later time points of this experiment? You would also expect the spots to be red, because cells that overexpress this transcription factor will have an increased rate of transcription, so it is induced and we see red spots. This is because Yap1p allows for transcription in environmental distress, and glucose depletion would be a state of environmental distress that would signal Yap1p to initiate transcription.
- (Question 16, p. 121) Using the microarray data, how could you verify that you had truly deleted TUP1 or overexpressed YAP1 in the experiments described in questions 8 and 9? For genes that are glucose-repressed, if TUP1 has been deleted their specific placement on the DNA chip should always show a red spot, and most other spots should be yellow. This is because whether or not glucose is in the environment, the genes will still be expressed as they are not able to repressed by glucose in either a normal glucose or depleted glucose environment. For YAP1, for genes that are transcriptionally activated by YAP1 there should be red spots on the chip, and again mostly yellow spots for other genes. These red spots will occur later as these genes are not induced until they are in environmental distress, which signals Yap1p to act as a transcription initiator. Additionally, an overexpression of YAP1 should result in there being a red spot at the YAP1 gene placement, as if there is an overexpression of the factor protein that means its own gene expression has been induced.
Acknowledgements
I worked with my homework partner User:Hivanson. We texted one time. We worked on the first question together.
Except for what is noted above, this individual journal entry was completed by me and not copied from another source.
Kmill104 (talk) 18:02, 14 February 2024 (PST)
References
- Alberts et al. (2002) Molecular Biology of the Cell, Ch. 8: Microarrays
- Brown, P.O. & Botstein, D. (1999) Exploring the new world of the genome with DNA microarrays Nature Genetics 21: 33-37. https://doi.org/10.1038/4462
- Campbell, A.M. and Heyer, L.J. (2003), “Chapter 4: Basic Research with DNA Microarrays”, in Discovering Genomics, Proteomics, and Bioinformatics, Cold Spring Harbor Laboratory Press, pp. 107-124. (Available on Brightspace)
- DeRisi, J.L., Iyer, V.R., and Brown, P.O. (1997) Exploring the Metabolic and Genetic Control of Gene Expression on a Genomic Scale. Science 278: 680-686. DOI: https://doi.org/10.1126/science.278.5338.680
- LMU BioDB 2024. (2024). Week 5. Retrieved February 14, 2024, from https://xmlpipedb.cs.lmu.edu/biodb/spring2024/index.php/Week_5
User Page
Assignment Pages
- Week 1
- Week 2
- Week 3
- Week 4
- Week 5
- Week 6
- Week 8
- Week 9
- Week 10
- Week 11
- Week 12
- Week 13
- Week 14
- Week 15
Individual Journal Entry Pages
- Kmill104 Week 1
- Kmill104 Week 2
- MSymond1 KMill104 Week 3
- Monarch Initiative Week 4
- Kmill104 Week 5
- Kmill104 Week 6
- Kmill104 Week 8
- Kmill104 Week 9
- Kmill104 Week 10
- Kmill104 Week 11
- Kmill104 Week 12
- Data Analysts Week 13
- Data Analysts Week 14
- Data Analysts Week 15