Lena Week 8
From LMU BioDB 2013
Contents |
Part 1
- Media: Merrell_Compiled_Raw_Data_Vibrio_LH_20131015.txt
- Media: Merrell_Compiled_Raw_Data_Vibrio_LH_20131015.xls
- When Filtered p-value less than 0.05, 948 total results found.
Part 2
GenMAPP Expression Dataset Manager Procedure
- I used the 2009 version of the Vibrio cholerae Gene Database with GenMAPP.
- I loaded my txt file of Vibrio cholerae, and allowed Expression Dataset Manager to convert the data.
- The processed data was saved here. Media:Merrell_Compiled_Raw_Data_Vibrio_AJV.gex
- The exception file was saved here. Media:Merrell_Compiled_Raw_Data_Vibrio_AJV.EX.txt
- There were 772 errors due to genes not being found.
- I had more errors than my partner who was using the updated version, she has 121. I think the updated version probably had a more comprehensive list of genes for Vibrio cholerae.
- I customized the data set of Avg_LogFC_all with colors. Increased was set to pink and decreased was set to green. To create the criterion for increased I set the Fold Change to > 0.25 and the P Value to < 0.05. To set the criterion for decreased I set Fold Change to < -0.25 and P Value to < 0.05.
- The .gex file was saved here. Media:Merrell_Compiled_Raw_Data_Vibrio_AJV.gex
MAPPFinder Procedure
- My Partner and I worked on the increased criterion.
- From GenMAPP I launched MAPPFinder and opened the expression data set file listed above. I clicked on the "Increased" citeria, and checked boxes for "Gene Ontology" and "P Value." In the Browse box, I named the file Increased_LH_20131015-Criterion0-GO.
- I ran the MAPPFinder. The top 10 Gene Ontology terms were...
- Transport Activity
- Localization
- Macromolecule Metabolic Process
- Cellular Macromolecule Metabolic Process
- Cellular Bipolymer Biosnythetic Process
- Bipolymer Biosnythetic Process
- Bipolymer Metabolic Process
- Amino Acid Metabloic Process
- Cellular Amino Acid and Derivative Metabolic Process
- Cellular Nitrogen Compound Metabolic Process
- Here is an image of the results.
- We had very different lists of top 10 GO terms. Her's is probably more accurate since it is the updated version.
- I searched GO terms associated with the genes mentioned by Merrel et al.
- VC0028: metal ion binding, iron-sulfur cluster binding, 4 iron, 4 sulfur cluster binidng, catalytic activity
- VC0941: transferase activity, glycine hydroxymethyltranserase activity, lygase activity, dihydroxy-acid dehydrase activity,
- VC0869: catalytic activity, ligase activity,phosphoriboseylformylglycinamide synthase activity
- VC0051: nucleotide binding, ATP binding, catalytic activity, lyase activity, carboxy-lyase activity, phosphoribosylaminoimidazole carboxylase activity,
- VC0647: nucleotidyltransferase activity, polyribonucleotide nucleotidyltransferase activity, transferase activity, 3'-5' exoribonuclease activity, RNA binding, mitochondrion
- VC2350: deoxyribose-phosphate aldolase activity, lyase activity, catalytic activity
- VCA0583: outer membrane-bounded periplasmic space
- My buddy and I had different terms associated with the genes. Since the last update, more terms were added for 2010.
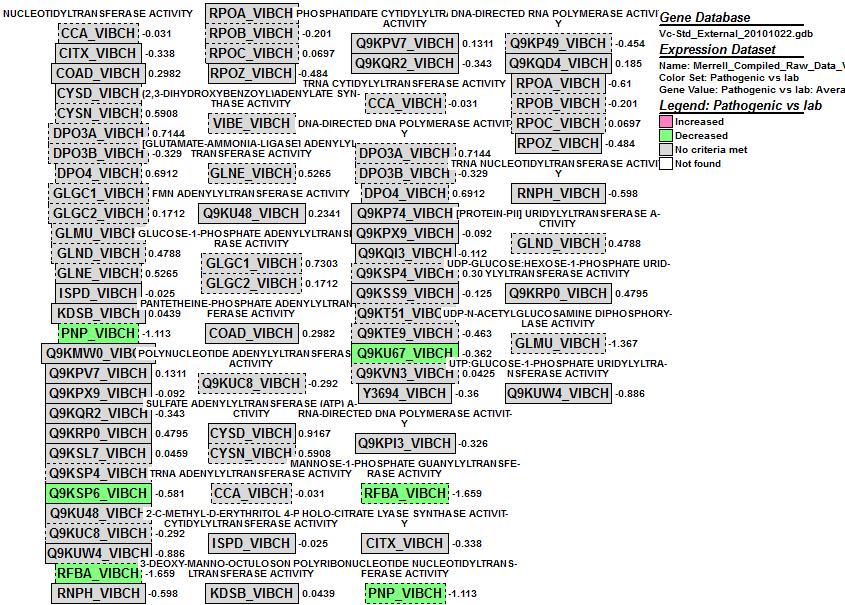
- I clicked on the GO term nucleotidyltransferase activity and this window came up. There were significant decreases in six places for VC0647.
Gene: PNP_VIBCH
- I doubled clicked on the gene PNP_VIBCH and was directed to this webpage. The function of this gene is to aid in mRNA degredation.
Uploaded Files
- Exception Files: Media:Merrell_Compiled_Raw_Data_Vibrio_AJV.EX.txt
- Expression Dataset file: Media:Merrell_Compiled_Raw_Data_Vibrio_AJV.gex
- GO Results file: Media:Increased_LH_20131015-Criterion0-GO.txt
- Go Results as Excel: Media:Increased_LH_20131015-Criterion0-GO.xls
- MAPP: not an uploadable file type, see screen shot above
- The MAPPFinder GO mappings file: Media:Merrell_Compiled_Raw_Data_Vibrio_AJV.gmf
Created by Lena Hunt