Nanguiano Week 7
From LMU BioDB 2015
Contents
Introduction to DNA Microarrays
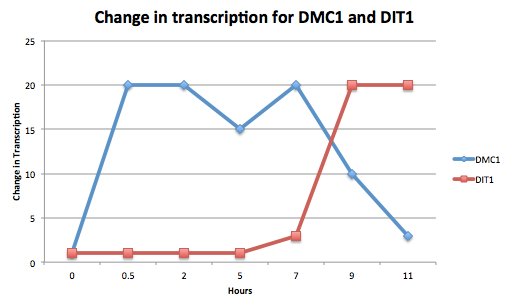
- (Question 5, p. 110) Choose two genes from Figure 4.6b (PDF of figures on MyLMUConnect) and draw a graph to represent the change in transcription over time. You can either create your plot in Excel and put the image up on your wiki page or you can do it in hard copy and turn it in in class.
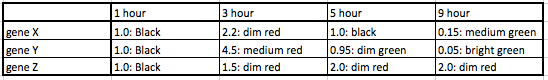
- (Question 6b, p. 110) Look at Figure 4.7, which depicts the loss of oxygen over time and the transcriptional response of three genes. These data are the ratios of transcription for genes X, Y, and Z during the depletion of oxygen. Using the color scale from Figure 4.6, determine the color for each ratio in Figure 4.7b. (Use the nomenclature "bright green", "medium green", "dim green", "black", "dim red", "medium red", or "bright red" for your answers.)
- (Question 7, p. 110) Were any of the genes in Figure 4.7b transcribed similarly? If so, which ones were transcribed similarly to which ones?
- Genes X and Y were transcribed similarly, though not the same. Both were upregulated at 3 hours, and downregulated at 9 hours. Y was very slightly downregulated at 5 hours, while X was neither up nor down-regulated at 5 hours.
- (Question 9, p. 118) Why would most spots be yellow at the first time point? I.e., what is the technical reason that spots show up as yellow - where does the yellow color come from? And, what would be the biological reason that the experiment resulted in most spots being yellow?
- The yellow color indicates that there has been no change in transcription. Most spots would be yellow at the first time point since conditions have not changed enough to warrant a change in transcription. The biological reason that the experiment resulted in most spots being yellow is that the experiment did not change conditions that resulted in a large change in transcription. The conditions may have caused changes in transcription in some pathways, but it was not widespread.
- (Question 10, p. 118) Go to the Saccharomyces Genome Database and search for the gene TEF4; you will see it is involved in translation. Look at the time point labeled OD 3.7 in Figure 4.12, and find the TEF4 spot. Over the course of this experiment, was TEF4 induced or repressed? Hypothesize why TEF4’s change in expression was part of the cell’s response to a reduction in available glucose (i.e., the only available food).
- Over the course of the experiment, TEF4 was repressed. TEF4's reduction in expression was part of the cell's response to the reduction in available glucose because translation requires energy. Reducing the amount of translation will reduce the amount of energy being used by the cell, thus allowing it to survive through starvation conditions longer than it would have otherwise.
- (Question, 11, p. 120) Why would TCA cycle genes be induced if the glucose supply is running out?
- TCA cycle genes would be induced because the TCA cycle increases the energy production of the cell. This allows to cell to produce and store the energy they would need to survive starvation while they still have a food source to consume.
- (Question 12, p. 120) What mechanism could the genome use to ensure genes for enzymes in a common pathway are induced or repressed simultaneously?
- The genome could ensure that the genes for enzymes in a common pathway are induced or repressed simultaneously by grouping the genes together in operons that are promoted or inhibited by the same transcription factor.
- (Question 13, p. 121) Consider a microarray experiment where cells deleted for the repressor TUP1 were subjected to the same experiment of a timecourse of glucose depletion where cells at t0 (plenty of glucose available) are labeled green and cells at later timepoints (glucose depleted) are labeled red. What color would you expect the spots that represented glucose-repressed genes to be in the later time points of this experiment?
- I would expect that the spots that represented glucose-repressed genes would be yellow, as the TUP1 repressor cannot repress them the way that they would be normally. Therefore, there is nothing to stop them from being transcribed as normal.
- (Question 14, p. 121) Consider a microarray experiment where cells that overexpress the transcription factor Yap1p were subjected to the same experiment of a timecourse of glucose depletion where cells at t0 (plenty of glucose available) are labeled green and cells at later timepoints (glucose depleted) are labeled red. What color would you expect the spots that represented Yap1p target genes to be in the later time points of this experiment?
- Yap1p increases the amount of expression in its target genes. As a result, if it is overexpressed, at the later timepoints, the Yap1p target genes would be bright red, as they would have been heavily expressed.
- (Question 16, p. 121) Using the microarray data, how could you verify that you had truly deleted TUP1 or overexpressed YAP1 in the experiments described in questions 8 and 9?
- You could verify that TUP1 had been deleted by watching the change in expression over time. If the dots remain yellow instead of turning green, then you know it has been deleted, as the expression is not changing the way it would if it was still present. To verify that YAP1 had been overexpressed, you can also watch the transcription over time. The spots should be bright red, indicating that they have been highly induced.
Links
Nicole Anguiano
BIOL 367, Fall 2015
Assignment Links
- Week 1 Assignment
- Week 2 Assignment
- Week 3 Assignment
- Week 4 Assignment
- Week 5 Assignment
- Week 6 Assignment
- Week 7 Assignment
- Week 8 Assignment
- Week 9 Assignment
- Week 10 Assignment
- Week 11 Assignment
- Week 12 Assignment
- Week 14 Assignment
- Week 15 Assignment
Individual Journals
- Individual Journal Week 2
- Individual Journal Week 3
- Individual Journal Week 4
- Individual Journal Week 5
- Individual Journal Week 6
- Individual Journal Week 7
- Individual Journal Week 8
- Individual Journal Week 9
- Individual Journal Week 10
- Individual Journal Week 11
- Individual Assessment
- Deliverables