Difference between revisions of "Kevin Wyllie Week 3"
From LMU BioDB 2015
(Just saving my progress in case of a connection timeout.) |
(Saving my progress. Will finish this assignment tomorrow.) |
||
| Line 75: | Line 75: | ||
** '''-2''' Nter- L K C R Q R N W R L G Y T R N M V L G N V S S S I L S S L A I V P I E I -Cter (shown in red) | ** '''-2''' Nter- L K C R Q R N W R L G Y T R N M V L G N V S S S I L S S L A I V P I E I -Cter (shown in red) | ||
** '''-3''' No polypeptide - first codon is STOP. (shown in blue) | ** '''-3''' No polypeptide - first codon is STOP. (shown in blue) | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ===XMLPipeDB Match Practice=== | ||
| + | |||
| + | |||
| + | [[Image:Kwscreenshot5.jpg|right|thumb]] | ||
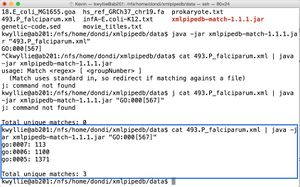
| + | * To count the occurrence of "GO:0005," "GO:0006," and "GO:0007" (shown on right): | ||
| + | |||
| + | cat 493.P_falciparum.xml | java -jar xmlpipedb-match-1.1.1.jar "GO:000[567]" | ||
| + | |||
| + | ** There are three unique matches (thus, each pattern did shown up). | ||
| + | ** "GO:0005" occurred 1,371 times. "GO:0006" occurred 1,100 times. "GO:0007" occurred 113 times. | ||
Revision as of 02:19, 20 September 2015
Contents
Complement of a Strand
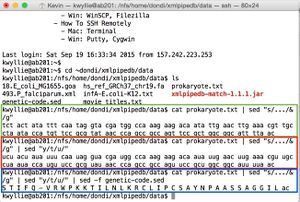
- Shown in green: the following command is used to open the file (using prokaryote.txt as an example).
cat prokaryote.txt
- Shown in red: the following command is used to sequence the complementary strand (in the 5' -> 3' direction - thus the "rev" command).
cat prokaryote.txt | sed "y/atgc/tacg/" | rev
- These commands yield the nucleotide sequence:
- 5'- gttaaaatgccgccagcggaactggcggctgggttatacgcggaacatggtattaggcaacgtttcaagttcaatattgtcttctttggccatcgtacctattgaaatatagtaga -3'
Reading Frames
The original sequence in the prokaryote.txt file will be assumed to be the top strand for this exercise.
+1, +2, and +3 Frames
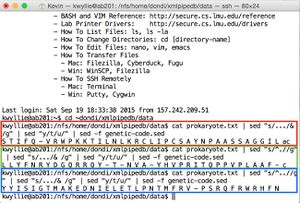
- Shown in green: to separate the strand into codons (resulting in the +1 frame):
cat prokaryote.txt | sed "s/.../& /g"
- Shown in red: to convert to the mRNA sequence (treating the DNA stand as the mRNA-like strand):
cat prokaryote.txt | sed "s/.../& /g" | sed "y/t/u/"
- Shown in blue: to translate this mRNA sequence (yielding the +1 frame):
cat prokaryote.txt | sed "s/.../& /g" | sed "y/t/u/" | sed -f genetic-code.sed
- For the +2 frame, the final pipe can be slightly altered:
cat prokaryote.txt | sed "s/^.//g" | sed "s/.../& /g" | sed "y/t/u/" | sed -f genetic-code.sed
- And similarly, for the +3 frame:
cat prokaryote.txt | sed "s/^..//g" | sed "s/.../& /g" | sed "y/t/u/" | sed -f genetic-code.sed
- These pipes yield the following amino acid sequences (shown on right):
- +1 Nter- S T I F Q -Cter (shown in green)
- +2 Nter- L L Y F N R Y D G Q R R Q Y -Cter (shown in red)
- +3 Nter- Y Y I S I G T M A K E D N I E L E T L P N T M F R V -Cter (shown in blue)
-1, -2, and -3 Frames
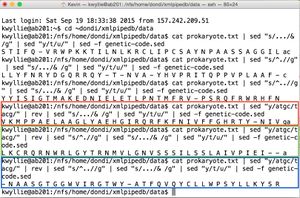
- For the -1 frame, open the file as usual, and then use the pipe from "Complement of a Strand" so that the commands after it will be applied to the complementary strand (instead of the original strand). Then, add the same pipe used for the +1 strand:
cat prokaryote.txt | sed "y/atgc/tacg/" | rev | sed "s/.../& /g" | sed "y/t/u/" | sed -f genetic-code.sed
- As before, the -2 and -3 frames can be found by making a single adjustment to the pipe for the -1 frame. For the -2 frame:
cat prokaryote.txt | sed "y/atgc/tacg/" | rev | sed "s/^.//g" | sed "s/.../& /g" | sed "y/t/u/" | sed -f genetic-code.sed
- And for the -3 frame:
cat prokaryote.txt | sed "y/atgc/tacg/" | rev | sed "s/^..//g" | sed "s/.../& /g" | sed "y/t/u/" | sed -f genetic-code.sed
- These pipes yield the following amino acid sequences (shown on right):
- -1 Nter- V K M P P A E L A A G L Y A E H G I R Q R F K F N I V F F G H R T Y -Cter (shown in green)
- -2 Nter- L K C R Q R N W R L G Y T R N M V L G N V S S S I L S S L A I V P I E I -Cter (shown in red)
- -3 No polypeptide - first codon is STOP. (shown in blue)
XMLPipeDB Match Practice
- To count the occurrence of "GO:0005," "GO:0006," and "GO:0007" (shown on right):
cat 493.P_falciparum.xml | java -jar xmlpipedb-match-1.1.1.jar "GO:000[567]"
- There are three unique matches (thus, each pattern did shown up).
- "GO:0005" occurred 1,371 times. "GO:0006" occurred 1,100 times. "GO:0007" occurred 113 times.