Kevin Wyllie Week 3
From LMU BioDB 2015
Contents
Complement of a Strand
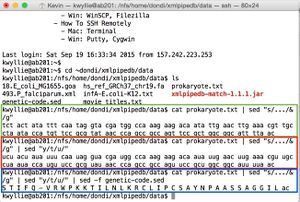
- Shown in green: the following command is used to open the file (using prokaryote.txt as an example).
cat prokaryote.txt
- Shown in red: the following command is used to sequence the complementary strand (in the 5' -> 3' direction - thus the "rev" command).
cat prokaryote.txt | sed "y/atgc/tacg/" | rev
- These commands yield the nucleotide sequence:
- 5'- gttaaaatgccgccagcggaactggcggctgggttatacgcggaacatggtattaggcaacgtttcaagttcaatattgtcttctttggccatcgtacctattgaaatatagtaga -3'
Reading Frames
The original sequence in the prokaryote.txt file will be assumed to be the top strand for this exercise.
+1, +2, and +3 Frames
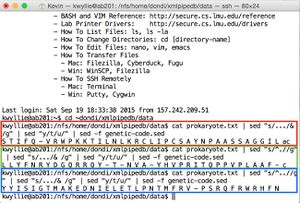
- Shown in green: to separate the strand into codons (resulting in the +1 frame):
cat prokaryote.txt | sed "s/.../& /g"
- Shown in red: to convert to the mRNA sequence (treating the DNA strand as the mRNA-like strand):
cat prokaryote.txt | sed "s/.../& /g" | sed "y/t/u/"
- Shown in blue: to translate this mRNA sequence (yielding the +1 frame):
cat prokaryote.txt | sed "s/.../& /g" | sed "y/t/u/" | sed -f genetic-code.sed
- For the +2 frame, the final pipe can be slightly altered:
cat prokaryote.txt | sed "s/^.//g" | sed "s/.../& /g" | sed "y/t/u/" | sed -f genetic-code.sed
- And similarly, for the +3 frame:
cat prokaryote.txt | sed "s/^..//g" | sed "s/.../& /g" | sed "y/t/u/" | sed -f genetic-code.sed
- These pipes yield the following amino acid sequences (shown on right):
- +1 Nter- S T I F Q - V R W P K K T I L N L K R C L I P C S A Y N P A A S S A G G I L -Cter (shown in red)
- +2 Nter- L L Y F N R Y D G Q R R Q Y - T - N V A - Y H V P R I T Q P P V P L A A F -Cter (shown in green)
- +3 Nter- Y Y I S I G T M A K E D N I E L E T L P N T M F R V - P S R Q F R W R H F N -Cter (shown in blue)
-1, -2, and -3 Frames
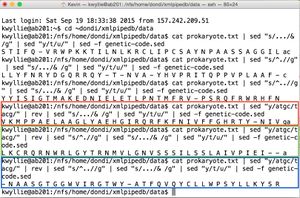
- For the -1 frame, open the file as usual, and then use the pipe from "Complement of a Strand" so that the commands after it will be applied to the complementary strand (instead of the original strand). Then, add the same pipe used for the +1 strand:
cat prokaryote.txt | sed "y/atgc/tacg/" | rev | sed "s/.../& /g" | sed "y/t/u/" | sed -f genetic-code.sed
- As before, the -2 and -3 frames can be found by making a single adjustment to the pipe for the -1 frame. For the -2 frame:
cat prokaryote.txt | sed "y/atgc/tacg/" | rev | sed "s/^.//g" | sed "s/.../& /g" | sed "y/t/u/" | sed -f genetic-code.sed
- And for the -3 frame:
cat prokaryote.txt | sed "y/atgc/tacg/" | rev | sed "s/^..//g" | sed "s/.../& /g" | sed "y/t/u/" | sed -f genetic-code.sed
- These pipes yield the following amino acid sequences (shown on right):
- -1 Nter- V K M P P A E L A A G L Y A E H G I R Q R F K F N I V F F G H R T Y - N I V -Cter (shown in red)
- -2 Nter- L K C R Q R N W R L G Y T R N M V L G N V S S S I L S S L A I V P I E I - - -Cter (shown in green)
- -3 Nter - - N A A S G T G G W V I R G T W Y - A T F Q V Q Y C L L W P S Y L L K Y S R - Cter. (shown in blue)
XMLPipeDB Match Practice
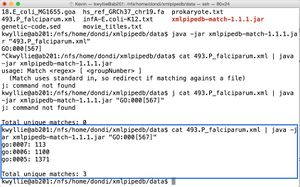
- To count the occurrence of GO:0005, GO:0006, and GO:0007 (shown on right):
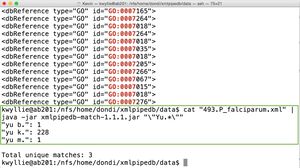
cat 493.P_falciparum.xml | java -jar xmlpipedb-match-1.1.1.jar "GO:000[567]"
- There are three unique matches (the maximum possible for this command).
- GO:0005 occurred 1,371 times.
- GO:0006 occurred 1,100 times.
- GO:0007 occurred 113 times.
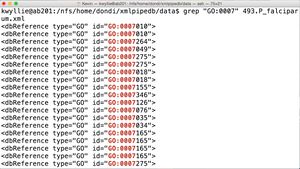
- To find GO:0007 "in situ" (shown on right):
grep "GO:0007" 493.P_falciparum.xml
- Looking at the text found on the same lines as this pattern, it appears to be the first few characters of a gene ID. Based on prior knowledge, it also may have something to do with gene ontology, as I have seen "GO" as an acronym for that term before.
- To count the occurrence of \"Yu.*\" (shown on right):
cat "493.P_falciparum.xml" | java -jar xmlpipedb-match-1.1.1.jar "\"Yu.*\""
- There are three unique matches.
- "yu b." occurred one time.
- "yu k." occurred 228 times.
- "yu m." occurred one time.
- A grep command for this pattern brings up lines such as:
<person name="Yu K."/>
So these may be names of biologists, perhaps those who were responsible for the discovery of a given gene.
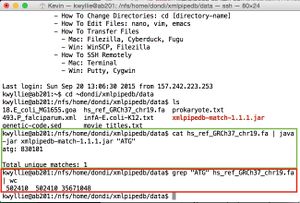
- To count occurrence of of "ATG."
- The match function finds 830,101 matches in hs_ref_GRCh37_chr19.fa (shown on right, in green).
- Connecting grep to wc finds 502,410 lines, 502,410 words and 35,671,048 characters (shown on right, in red).
- This discrepancy in matches is due to the differences in the functions. The Match function looks for the pattern outright, while grep-wc looks at the entirety of any line in which the pattern is found. The numbers that grep-wc returns apply to the lines that "ATG" is found in, not just the "ATG" pattern itself.
Protocol
Protocol - Complement of a Strand
- First, "ssh" into the server with the following command:
ssh <username>@my.cs.lmu.edu- The window will prompt you to enter your password. Type it in and press enter.
- Gain access to Dondi's folder with:
cd ~dondi/xmlpipedb/data - Open "prokaryote.txt" to view the DNA sequence it contains.
- To sequence the complementary strand, two operations must be done to the original DNA sequence.
- Each base of the original strand must be given its complement.
- The command that corresponds to this step is:
sed "y/atgc/tacg/". This replaces all G's with C's, all T's with A's, and so on.
- The command that corresponds to this step is:
- Since it is customary to express a nucleotide sequence in the 5' to 3' direction, the sequence must be reversed.
- The command that corresponds to this step is:
rev. Simply put, this reverses the sequence.
- The command that corresponds to this step is:
- Connecting these commands results in:
cat prokaryote.txt | sed "y/atgc/tacg/" | rev
- Each base of the original strand must be given its complement.
Protocol - Reading Frames
- To translate the +1 frame, three operations must be done on the original DNA sequence. Note: The following protocol treats the original strand as the mRNA like strand and the "top strand".
- The sequence must be separated into codons.
- The corresponding command is:
sed "s/.../& /g". This adds a space after every three characters, regardless of what those characters are.
- The corresponding command is:
- The T's must be changed to U's, since mRNA is the nucleotide sequence that gets translated, not DNA.
- The corresponding command is:
sed "y/t/u/". This changes all T's in the file to U's.
- The corresponding command is:
- Finally, the mRNA sequence must be translated.
- If done manually, an example of the many necessary commands would be:
sed "s/ATG/M/g". This would convert every ATG codon into an "M" for methionine, which is the amino acid that ATG codes for. - Fortunately, Dondi has graciously prepared a file that contains all of the necessary sed commands ("genetic-code.sed"). The syntax to apply this file's worth of commands is:
sed -f genetic-code.sed.
- If done manually, an example of the many necessary commands would be:
- Combining all of these commands results in the pipe:
cat prokaryote.txt | sed "s/.../& /g" | sed "y/t/u/" | sed -f genetic-code.sed.- Note: To end up with the correct amino acid sequence, it is required that the codons are separated by spaces before genetic-code.sed is applied. Otherwise, the first few sed commands in the file will translate between codons, disrupting the remaining codons.
- The sequence must be separated into codons.
- To translate the +2 frame, one additional operation must be done on the original DNA sequence.
- The first character in the sequence must be deleted (so that the frames are offset by one).
- The corresponding command is:
sed "s/^.//g". This "replaces" the first character in a sequence with nothing (effectively deleting it).
- The corresponding command is:
- Combining this command with the previous commands from the +1 frame results in the pipe:
cat prokaryote.txt | sed "s/^.//g" | sed "s/.../& /g" | sed "y/t/u/" | sed -f genetic-code.sed.- Note: To end up with the correct amino acid sequence, it is required that the first character is deleted before the codons are separated by spaces. Otherwise, the codons will effectively be the same as for the +1 frame, with the exception of the first codon failing to translate.
- The first character in the sequence must be deleted (so that the frames are offset by one).
- As with the +2 frame, another similar, additional step is required to translate the +3 frame.
- The first two characters in the sequence must be deleted (so that the frames are offset by one).
- The corresponding command is:
sed "s/^..//g". This "replaces" the first two characters in a sequence with nothing (effectively deleting them).
- The corresponding command is:
- Combining this command with the previous commands from the +1 frame results in the pipe:
cat prokaryote.txt | sed "s/^..//g" | sed "s/.../& /g" | sed "y/t/u/" | sed -f genetic-code.sed.- Note: To end up with the correct amino acid sequence, it is required that the first two characters are deleted before the codons are separated by spaces. Otherwise, the codons will effectively be the same as for the +1 frame, with the exception of the first codon failing to translate.
- The first two characters in the sequence must be deleted (so that the frames are offset by one).
- Frames -1, -2 and -3 can be translated similar to frames +1, +2 and +3, respectively, with two additional operations.
- Each base of the original strand must be given its complement.
- The corresponding command is:
sed "y/atgc/tacg/". This replaces all G's with C's, all T's with A's, and so on.
- The corresponding command is:
- Since polypeptides are expressed from the N-terminus to the C-terminus, the DNA sequence must be expressed in the 5' to 3' direction.
- The command that corresponds to this step is: rev . Simply put, this reverses the sequence.
- Placing these commands before either "plus" frame allows for the corresponding "minus" frame to be translated.
- For example, the command to translate the +2 frame is:
cat prokaryote.txt | sed "s/^.//g" | sed "s/.../& /g" | sed "y/t/u/" | sed -f genetic-code.sed. - Thus, the command to translate the -2 frame is:
cat prokaryote.txt | sed "y/atgc/tacg/" | rev | sed "s/^.//g" | sed "s/.../& /g" | sed "y/t/u/" | sed -f genetic-code.sed.
- For example, the command to translate the +2 frame is:
- Each base of the original strand must be given its complement.
Protocol - XMLPipeDB Match Practice
Protocol - Counting the Occurrence of a Pattern
- The XMLPipeDB Match file (xmlpipedb-match-1.1.1.jar) runs on java, and allows one to search a body of text for a given pattern.
- The given pattern
GO:000[567]is actually three patterns in one. The characters flanked by brackets, in Match's terms, are interchangeable. Thus, entering this pattern actually searches for "GO:0005," "GO:0006," and "GO:0007."
- The given pattern
- To execute the search (within the 493.P_falciparum.xml file), the following command is used:
cat 493.P_falciparum.xml | java -jar xmlpipedb-match-1.1.1.jar "GO:000[567]". - Match shows both the amount of times each individual pattern occurs, as well as the total number of "unique matches" that occurred at least once.
- For example, the maximum amount of unique matches for the pattern
GO:000[567]is three: one for each of the three effective patterns mentioned above.
- For example, the maximum amount of unique matches for the pattern
Protocol - Finding a Pattern "In Situ"
- The "grep" command is similar to Match in that it retrieves instances of a pattern within a file. However, grep returns the entire line in which the query is found.
- To find the pattern "GO:0007" within the 493.P_falciparum.xml file, use the command:
grep "GO:0007" 493.P_falciparum.xml. - Since grep also retrieves the text surrounding the pattern, this allows one to investigate into the meaning or purpose of a given pattern.
Protocol - Differentiating Between Match and Grep-wc
- The command "wc" allows for automated counting of the lines, words and characters in a file. The command for this is simply:
wc <file name>. - However, using the grep command, and connecting that command to wc will result in a word count for only the lines of text that grep retrieved. An example of a grep-wc command is:
grep "ATG" hs_ref_GRCh37_chr19.fa | wc. This will bring up all of the lines that the pattern "ATG" appears in, and amongst these lines, wc will count the lines, words and characters. - Thus, the primary difference between this string of commands and the Match function, is that Match looks for the pattern outright, while grep-wc will process all lines in which the given pattern is found.
Journal Links
- Kevin Wyllie Week 2 (See the original assignment and class journal.)
- Kevin Wyllie Week 3 (See the original assignment and class journal.)
- Kevin Wyllie Week 4 (See the original assignment and class journal.)
- Kevin Wyllie Week 5 (See the original assignment and class journal.)
- Kevin Wyllie Week 6 (See the original assignment and class journal.)
- Kevin Wyllie Week 7 (See the original assignment and class journal.)
- Kevin Wyllie Week 8 (See the original assignment and class journal.)
- Kevin Wyllie Week 9 (See the original assignment and class journal.)
- Kevin Wyllie Week 10 (See the original assignment.)
- Kevin Wyllie Week 11 (See the original assignment.)
- Kevin Wyllie Week 12 (See the original assignment.)
- Kevin Wyllie Week 14 (See the original assignment.)
- Kevin Wyllie Week 15 (See the original assignment.)