Kevin Wyllie Week 7
From LMU BioDB 2015
Questions
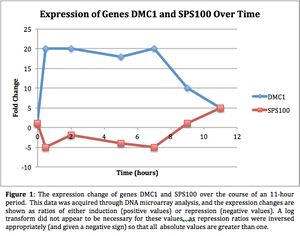
- (Question 5, p. 110) Choose two genes from Figure 4.6b and draw a graph to represent the change in transcription over time. You can either create your plot in Excel and put the image up on your wiki page or you can do it in hard copy and turn it in in class.
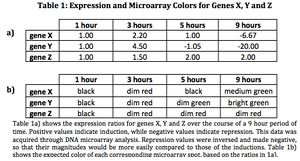
- (Question 6b, p. 110) Look at Figure 4.7, which depicts the loss of oxygen over time and the transcriptional response of three genes. These data are the ratios of transcription for genes X, Y, and Z during the depletion of oxygen. Using the color scale from Figure 4.6, determine the color for each ratio in Figure 4.7b. (Use the nomenclature "bright green", "medium green", "dim green", "black", "dim red", "medium red", or "bright red" for your answers.)
- (Question 7, p. 110) Were any of the genes in Figure 4.7b transcribed similarly? If so, which ones were transcribed similarly to which ones?
- Genes X and Y were transcribed similarly, in terms of induction versus repression. Both were induced between hours 1 and 2, went back down to approximately baseline expression by hour 5, and then saw significant repression between hours 5 and 9. However, in either repression or induction, gene Y saw a higher magnitude of regulation than gene X.
- (Question 9, p. 118) Why would most spots be yellow at the first time point? I.e., what is the technical reason that spots show up as yellow - where does the yellow color come from? And, what would be the biological reason that the experiment resulted in most spots being yellow?
- Because yellow signifies no (or very little) change in gene expression, it is not a surprise that, at the first time point in an experiment, most/many genes will show up as yellow on a microarray. It’s conceivable that the cell would require an initial startup period to detect and begin adjusting to an environmental condition. Gene regulatory mechanisms, though very small, are in fact composed of physical agents which operate in real time. As for the yellow color itself, it’s actually a bit of a visual misnomer. Microarray scanners don’t simply take a single colored picture of the slide. Rather, they detect each dye separately and then merge the images. Though yellow light may be the product of combining green and red light, yellow spots are artificially colored by the scanner’s software when it detects that an equal amount of Cy3 and Cy5 dyes were present in a given location on the slide.
- (Question 10, p. 118) Go to the Saccharomyces Genome Database and search for the gene TEF4; you will see it is involved in translation. Look at the time point labeled OD 3.7 in Figure 4.12, and find the TEF4 spot. Over the course of this experiment, was TEF4 induced or repressed? Hypothesize why TEF4’s change in expression was part of the cell’s response to a reduction in available glucose (i.e., the only available food).
- TEF4 appears to have been repressed in response to the reduction in glucose levels, as its spot has a slight green tint. The most intuitive explanation for this is because protein synthesis is very energetically costly, requiring multiple ATP equivalents to form a single peptide bond. In fact, in glucose deprivation, the cell will likely degrade proteins (to attain pyruvate from amino acid skeletons) rather than synthesize them. Not only will TEF4 be of little use in a cell that must reserve energy for more immediately-vital functions, but of course, Tef4p itself is a protein and thus would be wasteful to express in starvation conditions.
- (Question, 11, p. 120) Why would TCA cycle genes be induced if the glucose supply is running out?
- Ald2 and Acs1 would be induced because they allow for synthesis of acetyl-CoA, which is needed for the TCA cycle (the cell’s last resort now that glycolysis can’t occur). The ability of PEP carboxykinase and fructose-1,6-bisphosphatase to maintain higher glucose-6-phosphate levels is useful to the cell because G6P can be converted to pyruvate, which again, the cell would otherwise lack in the absence of glucose.
- (Question 12, p. 120) What mechanism could the genome use to ensure genes for enzymes in a common pathway are induced or repressed simultaneously?
- Operons are one thing that comes to mind, but I know that’s not the answer you’re looking for! A genome can regulate the expression of several genes in synchronicity by giving them the same/similar promoter regions, which will in turn be regulated by the same set of transcription factors. As Campbell and Heyer put it, “genes with similar expression profiles have similar promoters, and genes with similar promoters have similar expression profiles.”
- (Question 13, p. 121) Consider a microarray experiment where cells deleted for the repressor TUP1 were subjected to the same experiment of a timecourse of glucose depletion where cells at t0 (plenty of glucose available) are labeled green and cells at later timepoints (glucose depleted) are labeled red. What color would you expect the spots that represented glucose-repressed genes to be in the later time points of this experiment?
- Because Tup1p is important for the repression of glucose-repressed genes, genomes deleted for TUP1 will not be able to sense glucose. Effectively, they will always “think” that glucose is absent regardless of whether or not this is the case, and starvation genes will be expressed even when glucose is abundant. Thus, the spots of glucose-repressed genes will be yellow at any phase of the experiment. Though expression of glucose-repressed genes will be high, it will be equally high between different time points. Destroying the genome’s mechanism for detecting glucose will result in the same levels of expression both in glucose abundance and glucose deprivation.
- (Question 14, p. 121) Consider a microarray experiment where cells that overexpress the transcription factor Yap1p were subjected to the same experiment of a timecourse of glucose depletion where cells at t0 (plenty of glucose available) are labeled green and cells at later timepoints (glucose depleted) are labeled red. What color would you expect the spots that represented Yap1p target genes to be in the later time points of this experiment?
- Assuming glucose deprivation counts as an environmental stress that Yap1p responds to, overexpressing a transcription factor could have two different consequences. First, if Yap1p levels reach a certain, very high threshold, the protein may bind frequently enough to elicit a stress response even in the presence of glucose. This case is similar to the previous question, in that the genome’s ability to sense glucose levels is effectively disabled, so expression levels won’t vary, and the corresponding microarray spot will be yellow. The second possibility is that overexpression of Yap1p will make the cell highly sensitive to glucose levels, if the aforementioned threshold isn’t reached. If this is the case, the later time points will see much higher levels of stress response (gene expression) and the microarray spots will be strikingly red.
- (Question 16, p. 121) Using the microarray data, how could you verify that you had truly deleted TUP1 or overexpressed YAP1 in the experiments described in questions 8 and 9?
- TUP1 and YAP1 are both genes themselves, from which mRNA is synthesized. So microarray could easily indicate a deleted gene: if both sets of cDNA (or perhaps aRNA…) come from putatively-deleted cells (like in the previously mentioned glucose-deprivation experiment), there should be a complete absence of signal at the deleted gene’s microarray spot (no yellow, no green, no red). Conversely, to verify overexpression, the overexpressed transcriptome should be hybridized against that of a wild-type variant, which would result in a strikingly red microarray spot for the overexpressed gene.
Links
- Kevin Wyllie Week 2 (See the original assignment and class journal.)
- Kevin Wyllie Week 3 (See the original assignment and class journal.)
- Kevin Wyllie Week 4 (See the original assignment and class journal.)
- Kevin Wyllie Week 5 (See the original assignment and class journal.)
- Kevin Wyllie Week 6 (See the original assignment and class journal.)
- Kevin Wyllie Week 7 (See the original assignment and class journal.)
- Kevin Wyllie Week 8 (See the original assignment and class journal.)
- Kevin Wyllie Week 9 (See the original assignment and class journal.)
- Kevin Wyllie Week 10 (See the original assignment.)
- Kevin Wyllie Week 11 (See the original assignment.)
- Kevin Wyllie Week 12 (See the original assignment.)
- Kevin Wyllie Week 14 (See the original assignment.)
- Kevin Wyllie Week 15 (See the original assignment.)