Kevin Wyllie Week 3
From LMU BioDB 2015
Revision as of 19:15, 21 September 2015 by Kwyllie (Talk | contribs) (Saving my progress on the protocol.)
Contents
Complement of a Strand
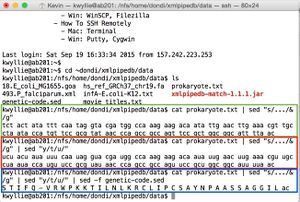
- Shown in green: the following command is used to open the file (using prokaryote.txt as an example).
cat prokaryote.txt
- Shown in red: the following command is used to sequence the complementary strand (in the 5' -> 3' direction - thus the "rev" command).
cat prokaryote.txt | sed "y/atgc/tacg/" | rev
- These commands yield the nucleotide sequence:
- 5'- gttaaaatgccgccagcggaactggcggctgggttatacgcggaacatggtattaggcaacgtttcaagttcaatattgtcttctttggccatcgtacctattgaaatatagtaga -3'
Reading Frames
The original sequence in the prokaryote.txt file will be assumed to be the top strand for this exercise.
+1, +2, and +3 Frames
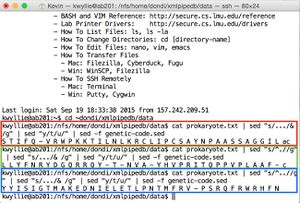
- Shown in green: to separate the strand into codons (resulting in the +1 frame):
cat prokaryote.txt | sed "s/.../& /g"
- Shown in red: to convert to the mRNA sequence (treating the DNA strand as the mRNA-like strand):
cat prokaryote.txt | sed "s/.../& /g" | sed "y/t/u/"
- Shown in blue: to translate this mRNA sequence (yielding the +1 frame):
cat prokaryote.txt | sed "s/.../& /g" | sed "y/t/u/" | sed -f genetic-code.sed
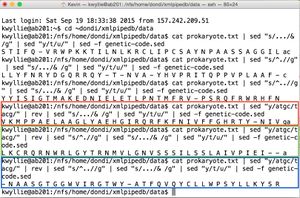
- For the +2 frame, the final pipe can be slightly altered:
cat prokaryote.txt | sed "s/^.//g" | sed "s/.../& /g" | sed "y/t/u/" | sed -f genetic-code.sed
- And similarly, for the +3 frame:
cat prokaryote.txt | sed "s/^..//g" | sed "s/.../& /g" | sed "y/t/u/" | sed -f genetic-code.sed
- These pipes yield the following amino acid sequences (shown on right):
- +1 Nter- S T I F Q - V R W P K K T I L N L K R C L I P C S A Y N P A A S S A G G I L -Cter (shown in red)
- +2 Nter- L L Y F N R Y D G Q R R Q Y - T - N V A - Y H V P R I T Q P P V P L A A F -Cter (shown in green)
- +3 Nter- Y Y I S I G T M A K E D N I E L E T L P N T M F R V - P S R Q F R W R H F N -Cter (shown in blue)
-1, -2, and -3 Frames
- For the -1 frame, open the file as usual, and then use the pipe from "Complement of a Strand" so that the commands after it will be applied to the complementary strand (instead of the original strand). Then, add the same pipe used for the +1 strand:
cat prokaryote.txt | sed "y/atgc/tacg/" | rev | sed "s/.../& /g" | sed "y/t/u/" | sed -f genetic-code.sed
- As before, the -2 and -3 frames can be found by making a single adjustment to the pipe for the -1 frame. For the -2 frame:
cat prokaryote.txt | sed "y/atgc/tacg/" | rev | sed "s/^.//g" | sed "s/.../& /g" | sed "y/t/u/" | sed -f genetic-code.sed
- And for the -3 frame:
cat prokaryote.txt | sed "y/atgc/tacg/" | rev | sed "s/^..//g" | sed "s/.../& /g" | sed "y/t/u/" | sed -f genetic-code.sed
- These pipes yield the following amino acid sequences (shown on right):
- -1 Nter- V K M P P A E L A A G L Y A E H G I R Q R F K F N I V F F G H R T Y - N I V -Cter (shown in red)
- -2 Nter- L K C R Q R N W R L G Y T R N M V L G N V S S S I L S S L A I V P I E I - - -Cter (shown in green)
- -3 Nter - - N A A S G T G G W V I R G T W Y - A T F Q V Q Y C L L W P S Y L L K Y S R - Cter. (shown in blue)
XMLPipeDB Match Practice
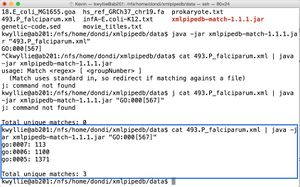
- To count the occurrence of GO:0005, GO:0006, and GO:0007 (shown on right):
cat 493.P_falciparum.xml | java -jar xmlpipedb-match-1.1.1.jar "GO:000[567]"
- There are three unique matches (the maximum possible for this command).
- GO:0005 occurred 1,371 times.
- GO:0006 occurred 1,100 times.
- GO:0007 occurred 113 times.
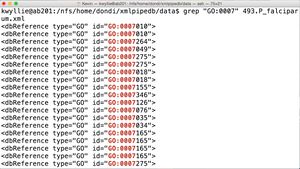
- To find GO:0007 "in situ" (shown on right):
grep "GO:0007" 493.P_falciparum.xml
- Looking at the text found on the same lines as this pattern, it appears to be the first few characters of a gene ID. Based on prior knowledge, it also may have something to do with gene ontology, as I have seen "GO" as an acronym for that term before.
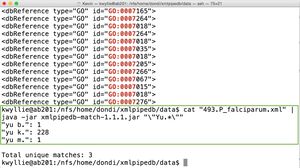
- To count the occurrence of \"Yu.*\" (shown on right):
cat "493.P_falciparum.xml" | java -jar xmlpipedb-match-1.1.1.jar "\"Yu.*\""
- There are three unique matches.
- "yu b." occurred one time.
- "yu k." occurred 228 times.
- "yu m." occurred one time.
- A grep command for this pattern brings up lines such as:
<person name="Yu K."/>
So these may be names of biologists, perhaps those who were responsible for the discovery of a given gene.
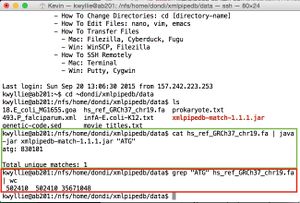
- To count occurrence of of "ATG."
- The match function finds 830,101 matches in hs_ref_GRCh37_chr19.fa (shown on right, in green).
- Connecting grep to wc finds 502,410 lines, 502,410 words and 35,671,048 characters (shown on right, in red).
- This discrepancy in matches is due to the differences in the functions. The Match function looks for the pattern outright, while grep-wc looks at the entirety of any line in which the pattern is found. The numbers that grep-wc returns apply to the lines that "ATG" is found in, not just the "ATG" pattern itself.
Protocol
Protocol - Complement of a Strand
- First, "ssh" into the server with the following command:
ssh <username>@my.cs.lmu.edu- The window will prompt you to enter your password. Type it in and press enter.
- Gain access to Dondi's folder with:
cd ~dondi/xmlpipedb/data - Open "prokaryote.txt" to view the DNA sequence it contains.
- To sequence the complementary strand, two operations must be done to the original DNA sequence.
- Each base of the original strand must be given its complement.
- The command that corresponds to this step is:
sed "y/atgc/tacg/". This replaces all G's with C's, all T's with A's, and so on.
- The command that corresponds to this step is:
- Since it is customary to express a nucleotide sequence in the 5' to 3' direction, the sequence must be reversed.
- The command that corresponds to this step is:
rev. Simply put, this reverses the sequence.
- The command that corresponds to this step is:
- Connecting these commands results in:
cat prokaryote.txt | sed "y/atgc/tacg/" | rev
- Each base of the original strand must be given its complement.
Protocol - Reading Frames
- To translate the +1 frame, three operations must be done on the original DNA sequence. Note: The following protocol treats the original strand as the mRNA like strand and the "top strand".
- The sequence must be separated into codons.
- The corresponding command is:
sed "s/.../& /g". This adds a space after every three characters, regardless of what those characters are.
- The corresponding command is:
- The T's must be changed to U's, since mRNA is the nucleotide sequence that gets translated, not DNA.
- The corresponding command is:
sed "y/t/u/". This changes all T's in the file to U's.
- The corresponding command is:
- Finally, the mRNA sequence must be translated.
- If done manually, an example of the many necessary commands would be:
sed "s/ATG/M/g". This would convert every ATG codon into an "M" for methionine, which is the amino acid that ATG codes for. - Fortunately, Dondi has graciously prepared a file that contains all of the necessary sed commands ("genetic-code.sed"). The syntax to apply this file's worth of commands is:
sed -f genetic-code.sed.
- If done manually, an example of the many necessary commands would be:
- The sequence must be separated into codons.