Blitvak Week 14
Contents
Goals for Week 14
- Consult with Anu to make modifications to TallyEngine/GenMAPP (share initial export results)
- Use Excel to track discrepant IDs (reference: Using Microsoft Excel to Compare ID Lists)
- Conduct gene database exports for any modified versions of GenMAPP builder that are created
- Analyze any conducted exports and perform Q&A work
Initial Export Analysis
Overview of Week 12 findings
- Using XMLPipeDB Match, 7127 unique matches were found that correlated with the OrderedLocusNames IDs outlined at the end of the week 12 assignment
- TallyEngine reported that 337 OrderedLocusNames were present in the XML and within the PSQL database
- Using
select count(*) from genenametype where type = 'ordered locus' and value ~ 'p?BCA[A-Z]?[0-9][0-9][0-9][A-Z]?[0-9]?[A-Z, a-z]?';, it was verified that 337 OrderedLocusNames entries were present in B.cenocepacia_J2315_20151119_gmb3build5. - By looking at the data present in the genenametype table, it was found that the OrderedLocusName data was in the format of
BceJ2315_#####
Steps taken for further analysis, conducted on 12/1
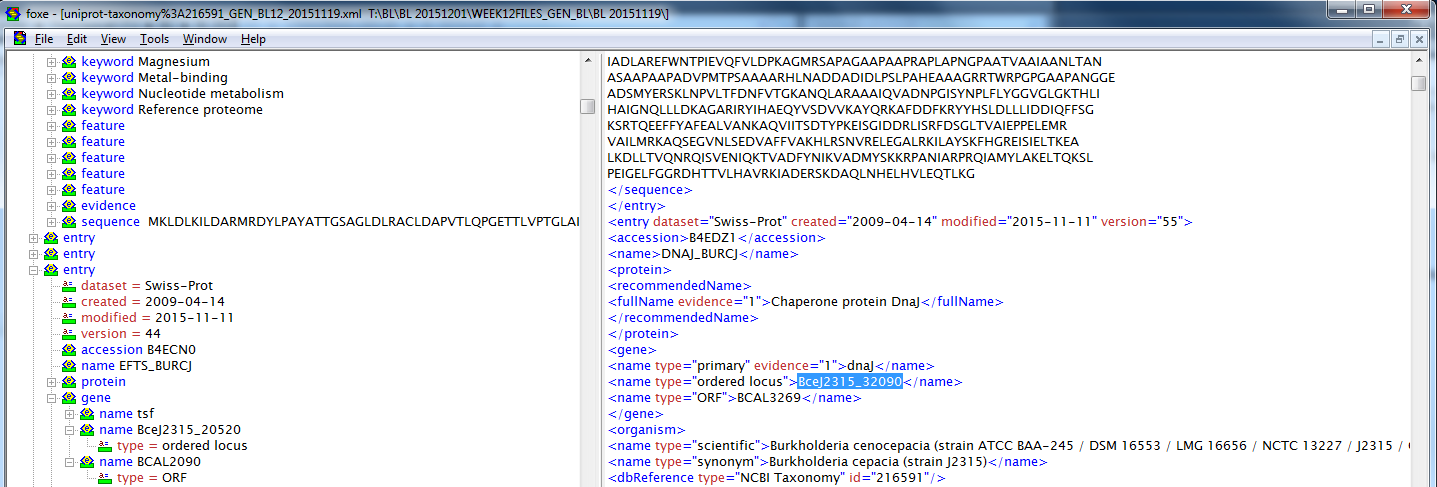
- The UniProt XML file was opened , via first object XML editor, in order to investigate and verify the location/nature of the OrderedLocusName data.
- A data entry was selected and the related data was looked into:
- In this entry, and in numerous others, it was noticed that only the gene name in the format of
BceJ2315_#####was tagged as being of the "ordered locus" type. The format that was being focused upon in previous work, that ofp?BCA[M,S,L]###?[A,a]#[A-Z]?, was labeled as being of the type "ORF". It was noticed that all entires that contained an "ordered locus" gene name also contained an "ORF" name for the same gene; most entries, additionally, lacked an "ordered locus" name and only contained an "ORF" name. - GenMAPP builder, by default, is made to pick up and utilize the ordered locus data within the XML; it was realized that, with respect to the initial export, it was functioning properly. Since the XML data only contained 337 OrderedLocus names, only 337 made it to the database. Since 7127 matches were found, using XMLPipeDB Match, that correlated to an "ORF" name, it is assumed that most of the gene data is ignored by focusing on OrderedLocus names.
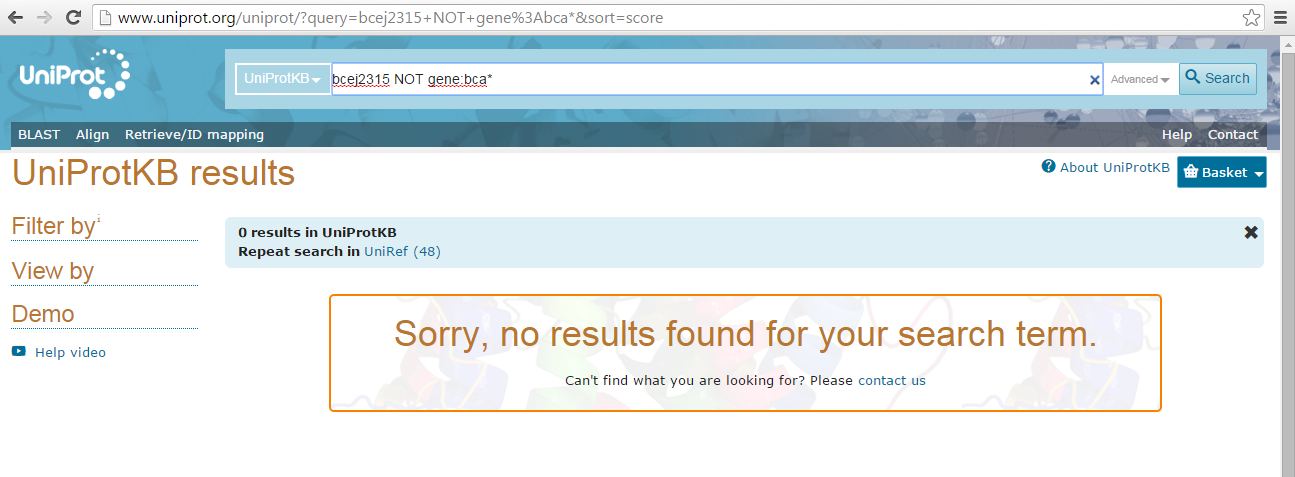
- UniProt KB was referenced in order to further verify that all
BceJ2315_#####gene names were coupled with one that was considered an "ORF" name - A search query was conducted that consisted of
bcej2315 NOT gene:bca*; this query, it was hoped, would show the number of gene entries that contained just an OrderedLocusName ID.
Discrepant ID analysis for the Initial Export, Conducted on 12/3
- The PSQL database for the initial export was opened up and the SQL query
select count(*) from genenametype where type = 'ORF' and value ~ 'p?BCA[A-Z]?[0-9][0-9][0-9][A-Z]?[0-9]?[A-Z, a-z]?';was run in order to find the ORF counts within the database (since TallyEngine, with the present used build of GenMAPP builder, did not incorporate the ORF data).- It was found that 7121 entries were within the database that corresponded to ORF data.
select * from genenametype where type = 'ORF';was run in order to observe the data; it was found that the data within the table corresponded to the gene name format of interest (p?BCA[A-Z]?[0-9][0-9][0-9][A-Z]?[0-9]?[A-Z, a-z]?).
- It was found that 7121 entries were within the database that corresponded to ORF data.
- However, it was found that there was a difference in count between what was previously reported by XMLPipeDB Match and by Postgres (7127 vs. 7121, a difference of 6 entries). It was realized that Excel should be utilized in order to track down the discrepant IDs.
Using Excel to track down discrepant IDs
- Pg Admin III was initialized and the database that was the initial export was booted up.
- The SQL query
select * from genenametype where type = 'ORF' order by valuewas utilized in order to put the data in ascending order (lower ID #s come first); the results of query was then exported in a format that Excel can read (text file). - Using the windows command line, through XMLPipeDB match, the 7127 unique XML entries that fit the criteria of ORF gene name were exported as a text file using
java -jar xmlpipedb-match-1.1.1.jar "p?BCA[A-Z]?[0-9][0-9][0-9][A-Z]?[0-9]?[A-Z, a-z]?" < uniprot-taxonomy%3A216591_GEN_BL12_20151119.xml > MATCHIDs_GEN_BL14_20151203 - Both files were opened with Excel and the proper settings were selected so that the gene name data ended up on its own column (for the Match data, a colon was selected as the divider between columns; PSQL data was comma separated).
- The column of IDs from from the Match utility, and the one that was found in the PostgreSQL database, were put side by side in a new Excel document with no spaces between them; it was ensured that each column was in ascending order. The column of IDs from the Match utility was given the label of "MATCH IDs"; the one from the PSQL database was given the label of "IDs FROM postgreSQL.
- Two new columns were created to the right of the ID columns, one was given the label of "MATCH: 1 to 2", the other that of "MATCH: 2 to 1". The plan, at this point, was to utilize Excel MATCH commands in order to compare the two sets of IDs with eachother; it was hoped that these commands would indicate which IDs were present in one set but not in the other.
- MATCH commands were then written the 2 MATCH columns and applied to the entirety of each MATCH column; the basic format is
=MATCH(VALUE TO LOOK-UP, RANGE/COLUMN WHERE THE LOOKING-UP OF A VALUE TAKES PLACE, "MATCH TYPE" [0 in this case]). The purpose of these MATCH commands is to compare the two different ID lists (with each other)
*ALL MATCH COMMANDS: Format - Column Label :MATCH Command(in first "cell") MATCH:1 to 2 :=MATCH(A2, B$2:B$7122, 0)MATCH:2 to 1 :=MATCH(B2, A$2:A$7122, 0)
Note: In the analysis conducted, the IDs from XMLPipeDB Match were placed in column A, and the ones from the Postgres Database were placed in column B. Values of "#N/A" appear in instances where an ID in one was set was not found in another.
- The Find function was used in Excel (via control + F) and the value #N/A was searched for ("Look In:" was set to Values). 6 instances of #N/A were found, which coheres with the difference of 6 that was found between the Match utility results and those of PSQL.
- The discrepant IDs are: bca199f, bca5253f, bca636c, bcad837b, bcal0235a, and bcal0239a
- UniProt yielded 0 results for query which further suggests that the "ORF" gene name should be focused upon; all entries in UniProt contained an "ORF" name and all of the entries found in the Model Organism Database for B. cenocepacia utilized gene names in the format of
p?BCA[M,S,L]###?[A,a]#[A-Z]?.- It was decided that GenMAPP builder should be modified so that it focuses, solely, on the "ORF" names within the XML file.
12/1
- Goals: Consult with Anu to make modifications to TallyEngine/GenMAPP (share initial export results)
- Reacquaint with using Excel to track IDs/discrepant IDs (Using Microsoft Excel to Compare ID Lists)
- pgAdmin III work will be involved, in conjunction with Excel, for the new modified .gdb: Will use:
select value from genenametype where type = 'ordered locus';and will export (and use with Excel)
B.cenocepacia_J2315_20151201_gmbuilder-genialomics-20151201
ORF not ORDEREDLOCUS
- XML - 3.72 minutes
- OBO/XML - 5.25 minutes
- Processing: 3.91 minutes
- GOA - 0.04 minutes
EXPORT START: 10:27 PM
END: 2:49 AM
12/3
'p?BCA[A-Z]?[0-9][0-9][0-9][A-Z]?[0-9]?[A-Z, a-z]?';
select count(*) from genenametype where type = 'ordered locus' and value ~ 'p?BCA[M,S,L][0-9][0-9][0-9][A,a]?[0-9]?[A-Z, a-z]?';
MODIFIED: 'p?BCA[M,S,L]?[0-9][0-9][0-9][A,a]?[0-9]?[A-Z, a-z]?';
first object xml editor
6 IDs with problems
bca199f
select count(*) from genenametype where type = 'ORF' and value ~ 'p?BCA[L,M,S]?[0-9][0-9][0-9][A,a]?[0-9]?[A-Z, a-z]?';
- BUILD 3
- XML - 3.99 minutes
- OBO/XML- 5.77 minutes (1), 4.06 minutes (2)
- GOA - 0.05 minutes
START: 7:24 p END: 12:01 a
- BUILD 4
- XML - 3.46 minutes
- OBO/XML - 5.05 minutes, 3.75 minutes (2)
- GOA - 0.04 minutes
START: 7:51 a END: 6:57 p
'[pBCA,BCAL,BCAS,BCAM][0-9][0-9][0-9][A,a]?[0-9]?[A-Z, a-z]?';
'(pBCA)?(BCAL)?(BCAS)?(BCAM)?[0-9][0-9][0-9][A,a]?[0-9]?[A-Z, a-z]?';