Kevin Wyllie Week 3
From LMU BioDB 2015
Revision as of 00:54, 20 September 2015 by Kwyllie (Talk | contribs) (Just saving my progress in case of a connection timeout.)
Journal Week 3
Complement of a Strand
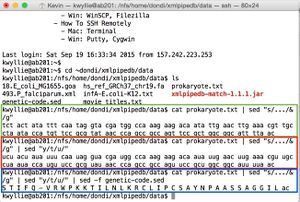
- Shown in green: the following command is used to open the file (using prokaryote.txt as an example).
cat prokaryote.txt
- Shown in red: the following command is used to sequence the complementary strand (in the 5' -> 3' direction - thus the "rev" command).
cat prokaryote.txt | sed "y/atgc/tacg/" | rev
- Finally, the sequence of the complementary strand is (copy/pasted from the command line window):
5'- gttaaaatgccgccagcggaactggcggctgggttatacgcggaacatggtattaggcaacgtttcaagttcaatattgtcttctttggccatcgtacctattgaaatatagtaga -3'
Reading Frames
The original sequence in the prokaryote.txt file will be assumed to be the top strand for this exercise.
- Shown in green: to separate the strand into codons (resulting in the +1 frame):
cat prokaryote.txt | sed "s/.../& /g"
- Shown in red: to convert to the mRNA sequence (treating the DNA stand as the mRNA-like strand):
cat prokaryote.txt | sed "s/.../& /g" | sed "y/t/u/"
- Shown in blue: to translate this mRNA sequence:
cat prokaryote.txt | sed "s/.../& /g" | sed "y/t/u/" | sed -f genetic-code.sed
- For the +2 frame, the final pipe can be slightly altered:
cat prokaryote.txt | sed "s/^.//g" | sed "s/.../& /g" | sed "y/t/u/" | sed -f genetic-code.sed
- And similarly, for the +3 frame:
cat prokaryote.txt | sed "s/^..//g" | sed "s/.../& /g" | sed "y/t/u/" | sed -f genetic-code.sed