Zvanysse Week 15
Contents
Notebook for Week 15
- As the teams were finishing up their code, we worked on major integrations from JASPAR, Gene Database APIs, and Page Design teams.
- It was difficult to communicate with certain teams on integrating, but once it was escalated it was resolved to a level in which we were able to complete our tasks on time.
- Specifically, we added NCBI, SGD, Ensemble, Uniprot, and JASPAR ID's to the top of the page.
- Through communication with Dondi,we found the correct ID format on how to reference the info.html and api.js file.
- Below is how the code looks:
var ncbiHrefTemplate = "https://www.ncbi.nlm.nih.gov/gene/";- Above is a template version for the genes information that comes from NCBI, now we can tag the unique gene to the template pathway as follows:
var ncbiId = gene.ncbi.ncbiID;- So the above code extends the pathway to find the ncbi ID for the specific gene.
$(".ncbi-link").text(ncbiId).attr({ href: ncbiHrefTemplate + ncbiId });- The "$" references the query api. Basically, this above line of code strings together the code adding the template and the specific part together.
- We had to link the data from the api.js file to where the page design team wanted the data in the info.html file
- We made each ID and built a skeleton so that once the api calls were all situated, it wouldn't be a problem once they lock down the correct path.
- Currently we have all the data being correctly pulled.
- For HMO1, for example, this is what the heading looks like:
- Once we saw this working, we took the majority of the code and replicated it for the other data fields
- We ran into trouble when trying to input the frequency matrix because the for the other page, we were using P-tags, but for this frequency matrix, we had to implement it another way.
- Initially we had some problems so we consulted our good friends Dr. Dionisio and the Eddies who helped us with the formatting of the table.
- because the length was not a set in stone length, we used a for loop to circumnavigate the unknown length. Find the code below:
- Frequency matrix code:
var frequencyMatrix = gene.jaspar.frequencyMatrix;var a = "";for (var i = 0; i < frequencyMatrix.A.length; i++) {a += "<td>" + frequencyMatrix.A[i] + "</td>";}$(".frequencyOfA").append($(a));
- We also had trouble with the actual implantation of the logo. We quickly found out that a src tag is supposed to be used to link between the API call thanks to Dondi. Eddie B. Also helped us with explaining how to retrieve the image URL.
- Specifically this can be seen here:
var sequenceLogo = gene.jaspar.sequenceLogo;$(".sequenceLogo").attr({ src : sequenceLogo });
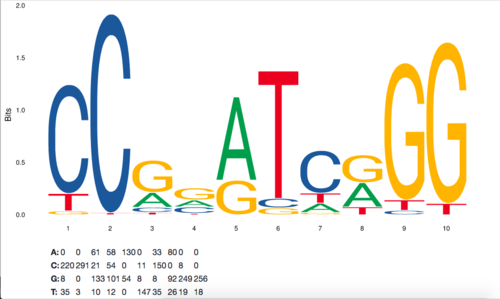
- This is an example of the Frequency Matrix and Sequence Logo for ASH1:
- Now that all the data has been linked and integrated, collaboration with the page design team and other biologists will help format the page to look aesthetically pleasing.
- For now, this is the current design for the HOT1 page:
Team Deliverable Progress
Overall, this week was primarily focused on wrapping up the code and finalizing the presentation. The presentation was practiced and rehearsed by all of us when we met in person. Katie focused heavily on the presentation aesthetic as well as making sure that she understood everything that was going on so that she would be able to help as needed. Emma finalized her data analysis portion. We all worked together on putting together the powerpoint presentation and made sure that each person had enough time to speak. As for the actual deliverables, the final pulls will be done before Friday 15th. If any major integration problems arise from last minute changes due to other teams, we will be on standby to make sure it is all worked out. The paper has been in the works with all of us contributing.
Acknowledgments
- I worked with Blair Hamilton to collaborate on the integration portion of our assignment. We met up in person as well as communicated through texting.
- We worked with Dr. Dionisio for figuring out the id Syntax and class references.
- We acknowledge the teams of JASPAR, Gene Database APIs, and Page Design throughout integrations.
- Collaborated with Eddie Bachoura on creating a table/for-loop for the frequency matrix portion of the info.html page. (thanks so much)
- Collaborated with Eddie Azinge on the gene name referencing between pages. For example, to get the gene name to appear at the top of the info.html page, Eddie A. helped by showing Blair and I how to attach an id to the name I wanted and then reference it in the info.js file.
- While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source.
Zvanysse (talk) 11:26, 13 December 2017 (PST)
References
LMU BioDB 2017. (2017). Deliverables. Retrieved November 14, 2017, from https://xmlpipedb.cs.lmu.edu/biodb/fall2017/index.php/GRNsight_Gene_Page_Project_Deliverables
LMU BioDB 2017. (2017). Week 15. Retrieved December 5, 2017, from https://xmlpipedb.cs.lmu.edu/biodb/fall2017/index.php/Week_15
BIOL/CMSI 367-01: Biological Databases Fall 2017
Assignments
Week 1 | Week 2 | Week 3 | Week 4 | Week 5 | Week 6 | Week 7 | Week 8 | Week 9 | Week 10 | Week 11 | Week 12 | Week 14
Individual Assignments
Zvanysse Week 1 | Zvanysse Week 2 | Zvanysse Week 3 | Zvanysse Week 4 | Zvanysse Week 5 | Zvanysse Week 6 | Zvanysse Week 7 | Zvanysse Week 8 | Zvanysse Week 9 | Zvanysse Week 10 | Zvanysse Week 11 | Zvanysse Week 12 | Zvanysse Week 14 | Zvanysse Week 15
Zvanysse Week 1 Journal | Zvanysse Week 2 Journal | Zvanysse Week 3 Journal | Zvanysse Week 4 Journal | Zvanysse Week 5 Journal | Zvanysse Week 6 Journal | Zvanysse Week 7 Journal | Zvanysse Week 8 Journal | Zvanysse Week 9 Journal | Zvanysse Week 10 Journal | Zvanysse Week 11 Journal | Zvanysse Week 12 Journal | Zvanysse Week 14 Journal