Difference between revisions of "Bhamilton18 Week 3"
From LMU BioDB 2017
Bhamilton18 (talk | contribs) (Changed picture sizing) |
Bhamilton18 (talk | contribs) (added category) |
||
| (12 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | ==Picture | + | ==Part I Picture Section== |
| − | + | ===Screenshots=== | |
| − | = | + | ====Basic LMU Website before Additions==== |
| − | |||
| − | ===Basic LMU Website before Additions=== | ||
[[file:Before Hacking Blair.png|1000px]] <!--Screeshots taken from www.lmu.edu website--> | [[file:Before Hacking Blair.png|1000px]] <!--Screeshots taken from www.lmu.edu website--> | ||
| Line 11: | Line 9: | ||
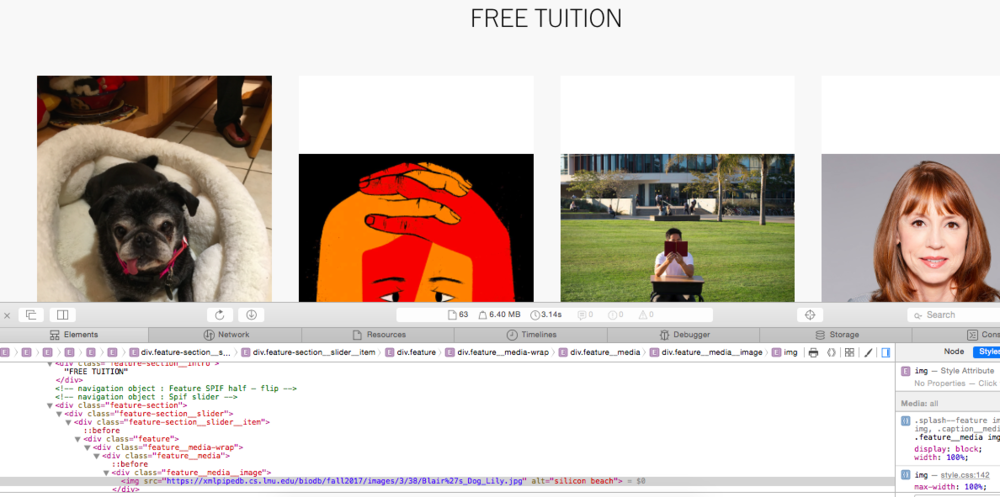
| − | ===After Additions with Inspected Elements=== | + | ====After Additions with Inspected Elements==== |
[[file:After Hacking LMU.png|1000px]] | [[file:After Hacking LMU.png|1000px]] | ||
| Line 17: | Line 15: | ||
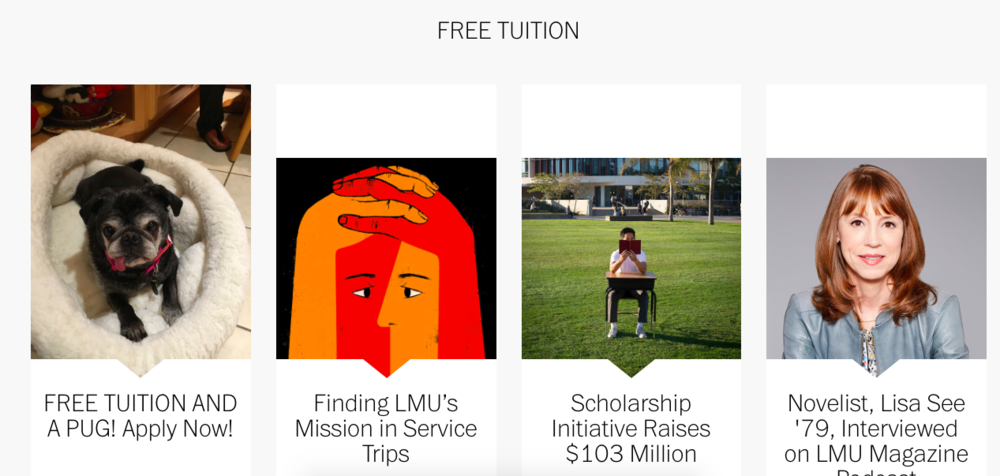
| − | ===Fake Website=== | + | ====Fake Website==== |
[[file:Apiril Fools Website.png|1000px]] | [[file:Apiril Fools Website.png|1000px]] | ||
| + | ==Part II Terminal Window Section== | ||
| + | |||
| + | ===Curling Raw Data=== | ||
| + | curl -d "pre_text=cgatggtacatagtagccgtagtgatgagatcgatgagctagc&submit=Submit" http://web.expasy.org/cgi-bin/translate/dna_aa <!-- (-d) represents the raw data selection. and the URL takes out the "action" section of the inspect element and allows the input to be delivered--> | ||
| + | <!--https://curl.haxx.se/docs/manual.html used this website to explain how to extract data--> | ||
| + | |||
| + | ===Questions Regarding ExPASy=== | ||
| + | #Are there any links to other pages within the ExPASy translation server’s responses? List them and state where they go. (of course, if you aren’t sure, you can always go there with a web browser or curl) | ||
| + | #* Yes, the following link from the ExPASy page: "sib.css", "bas.css", "sib_print.css", "ga.js", and "dna_aa". The css sites refer to the "look and feel" of the website, such as buttons, formatting or colors. The js file refers to the actions computed once a button is pressed or a page is loaded. Finally the dna_aa file is a separate file that holds the data for the translation and where the code is accessed to perform the translation.<!-- Once a translation has been submitted these are the URLs referenced--> | ||
| + | #Are there any identifiers in the ExPASy translation server’s responses? List them and state what you think they identify. | ||
| + | #*action="/cgi-bin/translate/dna_aa" The "cgi" portion represents an identifier of an action that needs to be performed once the translate button is pushed. | ||
| + | #*name="pre_text" The pre-text identifies where the text box is and what location text is found. | ||
| + | #*form method="POST" This tells the browser how to send data to the server. | ||
| + | |||
| + | ===Just the Answers=== | ||
| + | curl -d "pre_text=cgatggtacatggagtccagtagccgtagtgatgagatcgatgagctagc&submit=Submit" http://web.expasy.org/cgi-bin/translate/dna_aa | sed "1,47d" | sed "13,44d" | sed 's/<[^>]*>//g' | sed "2s/[A-Z]/& /g" | sed "4s/[A-Z]/& /g" | sed "6s/[A-Z]/& /g" | sed "8s/[A-Z]/& /g" | sed "10s/[A-Z]/& /g" | sed "12s/[A-Z]/& /g" | sed "s/-/STOP /g" | sed "s/M/Met/g" | ||
| + | |||
| + | |||
| + | ==Notebook== | ||
| + | |||
| + | #When using the "-d" function we are extracting the raw data from the file. We input a strand into the text bar through the command window and then retrieve every element of the URL including formatting, the translation and other parts of the web page. | ||
| + | #When looking up the Question answers, I collaborated with [[user:zvanysse| Zach Van Ysseldyk]] to inspect the resources on the page and decide which were referring to alienate pages and which were just "Google" markers. Next, I looked to find different "identifiers" on the web page, specifically filenames, paths and URLs. | ||
| + | #When looking to find just the answers, I first focused on getting rid of the more "extra" code. I began by deleting the text before and after the genetic code. Then I focused on getting rid of the "stuff" in between the brackets. After I looked to separate the capital letters from one another and add "STOP" codons and "Met" codons. | ||
| + | {{Template:Bhamilton18}} | ||
==Acknowledgements== | ==Acknowledgements== | ||
#This week I worked with my partner [[user:mbalducc|Mary Balducci]] on the hack-a-page segment of the homework. We collaborated on which website to "hack" and how to format the screenshots/pictures onto our respective web pages. | #This week I worked with my partner [[user:mbalducc|Mary Balducci]] on the hack-a-page segment of the homework. We collaborated on which website to "hack" and how to format the screenshots/pictures onto our respective web pages. | ||
#Also compared picture formatting to [[user:mbalducc|Mary Balducci's]] as it conveyed the correct title and formatting I wanted for my page. | #Also compared picture formatting to [[user:mbalducc|Mary Balducci's]] as it conveyed the correct title and formatting I wanted for my page. | ||
| + | #I worked with [[user:zvanysse| Zach Van Ysseldyk]] on the "curling" portion of the assignment. We collaborated on how the "curl" command works and what to look out for when computing the commands in the terminal. | ||
| + | #I reached out to [[user:dondi| Dr. Dioniso]] about different referencing criteria, as well as, what an identifier is and how to find them. | ||
| + | #I used the [http://www.lmu.edu| LMU Homepage] homepage in order to "hack" my dog and the text "FREE TUITION". | ||
#'''While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source.''' | #'''While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source.''' | ||
==References== | ==References== | ||
#LMU BioDB 2017. (2017). Week 3. Retrieved September 14, 2017, from https://xmlpipedb.cs.lmu.edu/biodb/fall2017/index.php/Week_3 | #LMU BioDB 2017. (2017). Week 3. Retrieved September 14, 2017, from https://xmlpipedb.cs.lmu.edu/biodb/fall2017/index.php/Week_3 | ||
| + | #Manual -- curl usage explained. Retrieved September 17, 2017, from https://curl.haxx.se/docs/manual.html | ||
| + | |||
| + | [[Category:Journal Entry]] | ||
Latest revision as of 04:29, 26 September 2017
Contents
Part I Picture Section
Screenshots
Basic LMU Website before Additions
After Additions with Inspected Elements
Fake Website
Part II Terminal Window Section
Curling Raw Data
curl -d "pre_text=cgatggtacatagtagccgtagtgatgagatcgatgagctagc&submit=Submit" http://web.expasy.org/cgi-bin/translate/dna_aa
Questions Regarding ExPASy
- Are there any links to other pages within the ExPASy translation server’s responses? List them and state where they go. (of course, if you aren’t sure, you can always go there with a web browser or curl)
- Yes, the following link from the ExPASy page: "sib.css", "bas.css", "sib_print.css", "ga.js", and "dna_aa". The css sites refer to the "look and feel" of the website, such as buttons, formatting or colors. The js file refers to the actions computed once a button is pressed or a page is loaded. Finally the dna_aa file is a separate file that holds the data for the translation and where the code is accessed to perform the translation.
- Are there any identifiers in the ExPASy translation server’s responses? List them and state what you think they identify.
- action="/cgi-bin/translate/dna_aa" The "cgi" portion represents an identifier of an action that needs to be performed once the translate button is pushed.
- name="pre_text" The pre-text identifies where the text box is and what location text is found.
- form method="POST" This tells the browser how to send data to the server.
Just the Answers
curl -d "pre_text=cgatggtacatggagtccagtagccgtagtgatgagatcgatgagctagc&submit=Submit" http://web.expasy.org/cgi-bin/translate/dna_aa | sed "1,47d" | sed "13,44d" | sed 's/<[^>]*>//g' | sed "2s/[A-Z]/& /g" | sed "4s/[A-Z]/& /g" | sed "6s/[A-Z]/& /g" | sed "8s/[A-Z]/& /g" | sed "10s/[A-Z]/& /g" | sed "12s/[A-Z]/& /g" | sed "s/-/STOP /g" | sed "s/M/Met/g"
Notebook
- When using the "-d" function we are extracting the raw data from the file. We input a strand into the text bar through the command window and then retrieve every element of the URL including formatting, the translation and other parts of the web page.
- When looking up the Question answers, I collaborated with Zach Van Ysseldyk to inspect the resources on the page and decide which were referring to alienate pages and which were just "Google" markers. Next, I looked to find different "identifiers" on the web page, specifically filenames, paths and URLs.
- When looking to find just the answers, I first focused on getting rid of the more "extra" code. I began by deleting the text before and after the genetic code. Then I focused on getting rid of the "stuff" in between the brackets. After I looked to separate the capital letters from one another and add "STOP" codons and "Met" codons.
Acknowledgements
- This week I worked with my partner Mary Balducci on the hack-a-page segment of the homework. We collaborated on which website to "hack" and how to format the screenshots/pictures onto our respective web pages.
- Also compared picture formatting to Mary Balducci's as it conveyed the correct title and formatting I wanted for my page.
- I worked with Zach Van Ysseldyk on the "curling" portion of the assignment. We collaborated on how the "curl" command works and what to look out for when computing the commands in the terminal.
- I reached out to Dr. Dioniso about different referencing criteria, as well as, what an identifier is and how to find them.
- I used the LMU Homepage homepage in order to "hack" my dog and the text "FREE TUITION".
- While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source.
References
- LMU BioDB 2017. (2017). Week 3. Retrieved September 14, 2017, from https://xmlpipedb.cs.lmu.edu/biodb/fall2017/index.php/Week_3
- Manual -- curl usage explained. Retrieved September 17, 2017, from https://curl.haxx.se/docs/manual.html