Difference between revisions of "Bhamilton18 Week 6"
From LMU BioDB 2017
Bhamilton18 (talk | contribs) (Changed some of my answers for #2) |
Bhamilton18 (talk | contribs) (answered final question and added reference) |
||
| (3 intermediate revisions by the same user not shown) | |||
| Line 11: | Line 11: | ||
#TEF4, or "translational elongation factor", was repressed, as the dot on the figure corresponded to bright green. The TEF4 could be reacting to the low amounts of glucose which provides energy to the cell. The TEF4 resorts to, according to Campbell, an energy saving mode therefore the genes were repressed. | #TEF4, or "translational elongation factor", was repressed, as the dot on the figure corresponded to bright green. The TEF4 could be reacting to the low amounts of glucose which provides energy to the cell. The TEF4 resorts to, according to Campbell, an energy saving mode therefore the genes were repressed. | ||
#During high stress environments genes are repressed and induced according to their energy levels. Due to TCA being a energy metabolic gene, it becomes induced in order to make energy efficiently and effectively. | #During high stress environments genes are repressed and induced according to their energy levels. Due to TCA being a energy metabolic gene, it becomes induced in order to make energy efficiently and effectively. | ||
| − | + | #The best mechanism a genome can use is a transcription factor to ensure genes for enzymes have a common induced or repressed pathway. | |
| − | + | #According to Campbell, TUP1 is responsible for the repression of glucose-repressed genes. Given this information I would expect the spots to be red, because the TUP1 and glucose would be depleted at said time period. | |
| − | + | #I would expect Yap1p to be colored red as it is "a transcription factor known to confer resistance to environmental stress"(121). During glucose depletion a cell in under stress, there Yap1p would increase expressed target genes and therefore be overly expressed and turn bright red. | |
| − | + | #To see if TUP1 has been deleted you can look at the change in time and see the coloring of the dot. If TUP! has been deleted it will turn black and not light up at all. To see if YAP1 has been overexpressed you can see once the gene turns bright red it is most likely highly induced. | |
{{Bhamilton18}} | {{Bhamilton18}} | ||
==Acknowledgments== | ==Acknowledgments== | ||
| − | #This week I worked with my partner[[User: Dbashour|Dina Bashoura]] on the discovery questions segment of the homework. We met in person as well as messaged each other if we were stuck on a question or needed more explanation. | + | #This week I worked with my partner [[User: Dbashour|Dina Bashoura]] on the discovery questions segment of the homework. We met in person as well as messaged each other if we were stuck on a question or needed more explanation. |
#'''While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source.''' | #'''While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source.''' | ||
==References== | ==References== | ||
| + | #Campbell, A.M. and Heyer, L.J. (2003), “Chapter 4: Basic Research with DNA Microarrays”, in Discovering Genomics, Proteomics, and Bioinformatics, Cold Spring Harbor Laboratory Press, pp. 107-124. | ||
#LMU BioDB 2017. (2017). Week 6. Retrieved October 5, 2017, from https://xmlpipedb.cs.lmu.edu/biodb/fall2017/index.php/Week_6#Individual_Journal_Assignment | #LMU BioDB 2017. (2017). Week 6. Retrieved October 5, 2017, from https://xmlpipedb.cs.lmu.edu/biodb/fall2017/index.php/Week_6#Individual_Journal_Assignment | ||
Latest revision as of 01:32, 10 October 2017
Discovery Questions
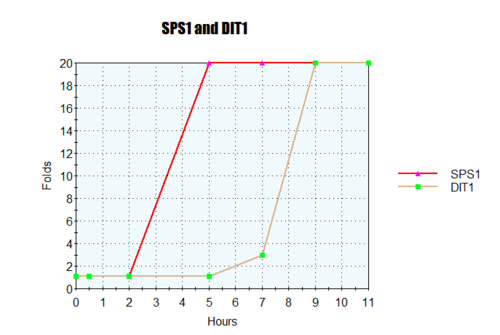
-
- Gene X: Hour 1: Black Hour 3: Dim Red Hour 5: Black Hour 9: Medium Green
- Gene Y: Hour 1: Black Hour 3: Medium Red Hour 5: Dim Green Hour 9: Bright Green
- Gene Z: Hour 1: Black Hour 3: Dim Red Hour 5: Dim Red Hour 9: Dim Red
- Gene X and Y were similar. Both had similar fold induction increase then deplete to 1:1 then proceed to fold repression.
-
- The technical reason these spots come up yellow is that there are equal amounts of green and red dye and when mixed they create yellow.
- The biological reason a majority were unchanged, i.e yellow, was because of the conditions that the experiment placed on the DNA slides.
- TEF4, or "translational elongation factor", was repressed, as the dot on the figure corresponded to bright green. The TEF4 could be reacting to the low amounts of glucose which provides energy to the cell. The TEF4 resorts to, according to Campbell, an energy saving mode therefore the genes were repressed.
- During high stress environments genes are repressed and induced according to their energy levels. Due to TCA being a energy metabolic gene, it becomes induced in order to make energy efficiently and effectively.
- The best mechanism a genome can use is a transcription factor to ensure genes for enzymes have a common induced or repressed pathway.
- According to Campbell, TUP1 is responsible for the repression of glucose-repressed genes. Given this information I would expect the spots to be red, because the TUP1 and glucose would be depleted at said time period.
- I would expect Yap1p to be colored red as it is "a transcription factor known to confer resistance to environmental stress"(121). During glucose depletion a cell in under stress, there Yap1p would increase expressed target genes and therefore be overly expressed and turn bright red.
- To see if TUP1 has been deleted you can look at the change in time and see the coloring of the dot. If TUP! has been deleted it will turn black and not light up at all. To see if YAP1 has been overexpressed you can see once the gene turns bright red it is most likely highly induced.
Acknowledgments
- This week I worked with my partner Dina Bashoura on the discovery questions segment of the homework. We met in person as well as messaged each other if we were stuck on a question or needed more explanation.
- While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source.
References
- Campbell, A.M. and Heyer, L.J. (2003), “Chapter 4: Basic Research with DNA Microarrays”, in Discovering Genomics, Proteomics, and Bioinformatics, Cold Spring Harbor Laboratory Press, pp. 107-124.
- LMU BioDB 2017. (2017). Week 6. Retrieved October 5, 2017, from https://xmlpipedb.cs.lmu.edu/biodb/fall2017/index.php/Week_6#Individual_Journal_Assignment