Bhamilton18 Week 6
From LMU BioDB 2017
Discovery Questions
-
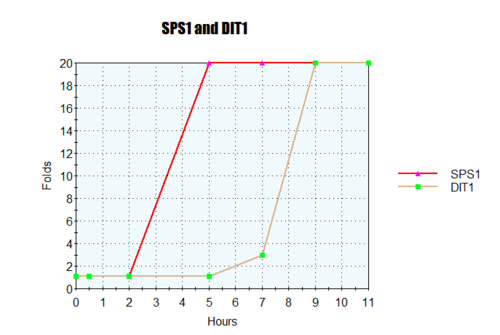
- Gene X: Hour 1: Black Hour 3: Dim Red Hour 5: Black Hour 9: Dim Green
- Gene Y: Hour 1: Black Hour 3: Medium Red Hour 5: Black Hour 9: Bright Green
- Gene Z: Hour 1: Black Hour 3: Black Hour 5: Dim Red Hour 9: Dim Red
- Gene X and Y were similar. Both had similar fold induction increase then deplete to 1:1 then proceed to fold repression.
-
- The technical reason these spots come up yellow is that there are equal amounts of green and red dye and when mixed they create yellow.
- The biological reason a majority were unchanged, i.e yellow, was because of the conditions that the experiment placed on the DNA slides.
- (Question 10, p. 118) Go to the Saccharomyces Genome Database and search for the gene TEF4; you will see it is involved in translation. Look at the time point labeled OD 3.7 in Figure 4.12, and find the TEF4 spot. Over the course of this experiment, was TEF4 induced or repressed? Hypothesize why TEF4’s change in expression was part of the cell’s response to a reduction in available glucose (i.e., the only available food).
- (Question, 11, p. 120) Why would TCA cycle genes be induced if the glucose supply is running out?
- (Question 12, p. 120) What mechanism could the genome use to ensure genes for enzymes in a common pathway are induced or repressed simultaneously?
- (Question 13, p. 121) Consider a microarray experiment where cells deleted for the repressor TUP1 were subjected to the same experiment of a timecourse of glucose depletion where cells at t0 (plenty of glucose available) are labeled green and cells at later timepoints (glucose depleted) are labeled red. What color would you expect the spots that represented glucose-repressed genes to be in the later time points of this experiment?
- (Question 14, p. 121) Consider a microarray experiment where cells that overexpress the transcription factor Yap1p were subjected to the same experiment of a timecourse of glucose depletion where cells at t0 (plenty of glucose available) are labeled green and cells at later timepoints (glucose depleted) are labeled red. What color would you expect the spots that represented Yap1p target genes to be in the later time points of this experiment?
- (Question 16, p. 121) Using the microarray data, how could you verify that you had truly deleted TUP1 or overexpressed YAP1 in the experiments described in questions 8 and 9?
Acknowledgments
- This week I worked with my partnerDina Bashoura on the discovery questions segment of the homework. We met in person as well as messaged each other if we were stuck on a question or needed more explanation.
- While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source.
References
- LMU BioDB 2017. (2017). Week 6. Retrieved October 5, 2017, from https://xmlpipedb.cs.lmu.edu/biodb/fall2017/index.php/Week_6#Individual_Journal_Assignment