Zvanysse Week 15
Contents
Notebook for Week 15
- As the teams were finishing up their code, we worked on major integrations from JASPAR, Gene Database APIs, and Page Design teams.
- It was difficult to communicate with certain teams on integrating, but once it was escalated it was resolved to a level in which we were able to complete our tasks on time.
- Specifically, we added NCBI, SGD, Ensemble, Uniprot, and JASPAR ID's to the top of the page.
- Through communication with Dondi,we found the correct ID format on how to reference the info.html and api.js file.
- Below is how the code looks:
var ncbiHrefTemplate = "https://www.ncbi.nlm.nih.gov/gene/";- Above is a template version for the genes information that comes from NCBI, now we can tag the unique gene to the template pathway as follows:
var ncbiId = gene.ncbi.ncbiID;- So the above code extends the pathway to find the ncbi ID for the specific gene.
$(".ncbi-link").text(ncbiId).attr({ href: ncbiHrefTemplate + ncbiId });- The "$" references the query api. Basically, this above line of code strings together the code adding the template and the specific part together.
- We had to link the data from the api.js file to where the page design team wanted the data in the info.html file
- We made each ID and built a skeleton so that once the api calls were all situated, it wouldn't be a problem once they lock down the correct path.
- Currently we have all the data being correctly pulled.
- For HMO1, for example, this is what the heading looks like:
- Once we saw this working, we took the majority of the code and replicated it for the other data fields
- We ran into trouble when trying to input the frequency matrix because the for the other page, we were using P-tags, but for this frequency matrix, we had to implement it another way.
- Initially we had some problems so we consulted our good friends Dr. Dionisio and the Eddies who helped us with the formatting of the table.
- because the length was not a set in stone length, we used a for loop to circumnavigate the unknown length. Find the code below:
- Frequency matrix code:
var frequencyMatrix = gene.jaspar.frequencyMatrix;var a = "";for (var i = 0; i < frequencyMatrix.A.length; i++) {a += "<td>" + frequencyMatrix.A[i] + "</td>";}$(".frequencyOfA").append($(a));
- We also had trouble with the actual implantation of the logo. We quickly found out that a src tag is supposed to be used to link between the API call thanks to Dondi. Eddie B. Also helped us with explaining how to retrieve the image URL.
- Specifically this can be seen here:
var sequenceLogo = gene.jaspar.sequenceLogo;$(".sequenceLogo").attr({ src : sequenceLogo });
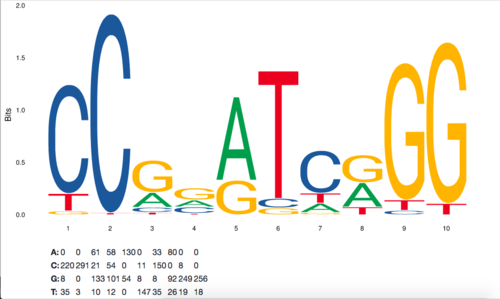
- This is an example of the Frequency Matrix and Sequence Logo for ASH1:
Acknowledgments
- I worked with Blair Hamilton to collaborate on the integration portion of our assignment. We met up in person as well as communicated through texting.
- We worked with Dr. Dionisio for figuring out the id Syntax and class references.
- We acknowledge the teams of JASPAR, Gene Database APIs, and Page Design throughout integrations.
- While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source.
Zvanysse (talk) 11:26, 13 December 2017 (PST)
References
LMU BioDB 2017. (2017). Deliverables. Retrieved November 14, 2017, from https://xmlpipedb.cs.lmu.edu/biodb/fall2017/index.php/GRNsight_Gene_Page_Project_Deliverables
LMU BioDB 2017. (2017). Week 15. Retrieved December 5, 2017, from https://xmlpipedb.cs.lmu.edu/biodb/fall2017/index.php/Week_15
BIOL/CMSI 367-01: Biological Databases Fall 2017
Assignments
Week 1 | Week 2 | Week 3 | Week 4 | Week 5 | Week 6 | Week 7 | Week 8 | Week 9 | Week 10 | Week 11 | Week 12 | Week 14
Individual Assignments
Zvanysse Week 1 | Zvanysse Week 2 | Zvanysse Week 3 | Zvanysse Week 4 | Zvanysse Week 5 | Zvanysse Week 6 | Zvanysse Week 7 | Zvanysse Week 8 | Zvanysse Week 9 | Zvanysse Week 10 | Zvanysse Week 11 | Zvanysse Week 12 | Zvanysse Week 14 | Zvanysse Week 15
Zvanysse Week 1 Journal | Zvanysse Week 2 Journal | Zvanysse Week 3 Journal | Zvanysse Week 4 Journal | Zvanysse Week 5 Journal | Zvanysse Week 6 Journal | Zvanysse Week 7 Journal | Zvanysse Week 8 Journal | Zvanysse Week 9 Journal | Zvanysse Week 10 Journal | Zvanysse Week 11 Journal | Zvanysse Week 12 Journal | Zvanysse Week 14 Journal