Knguye66 Week 6
Jump to navigation
Jump to search
Purpose
The purpose of this assignment is to read and learn about DNA microarray work and analyze its experiments from reading Chapter 4 of Campbell & Heyer (2003).
Questions & Answers
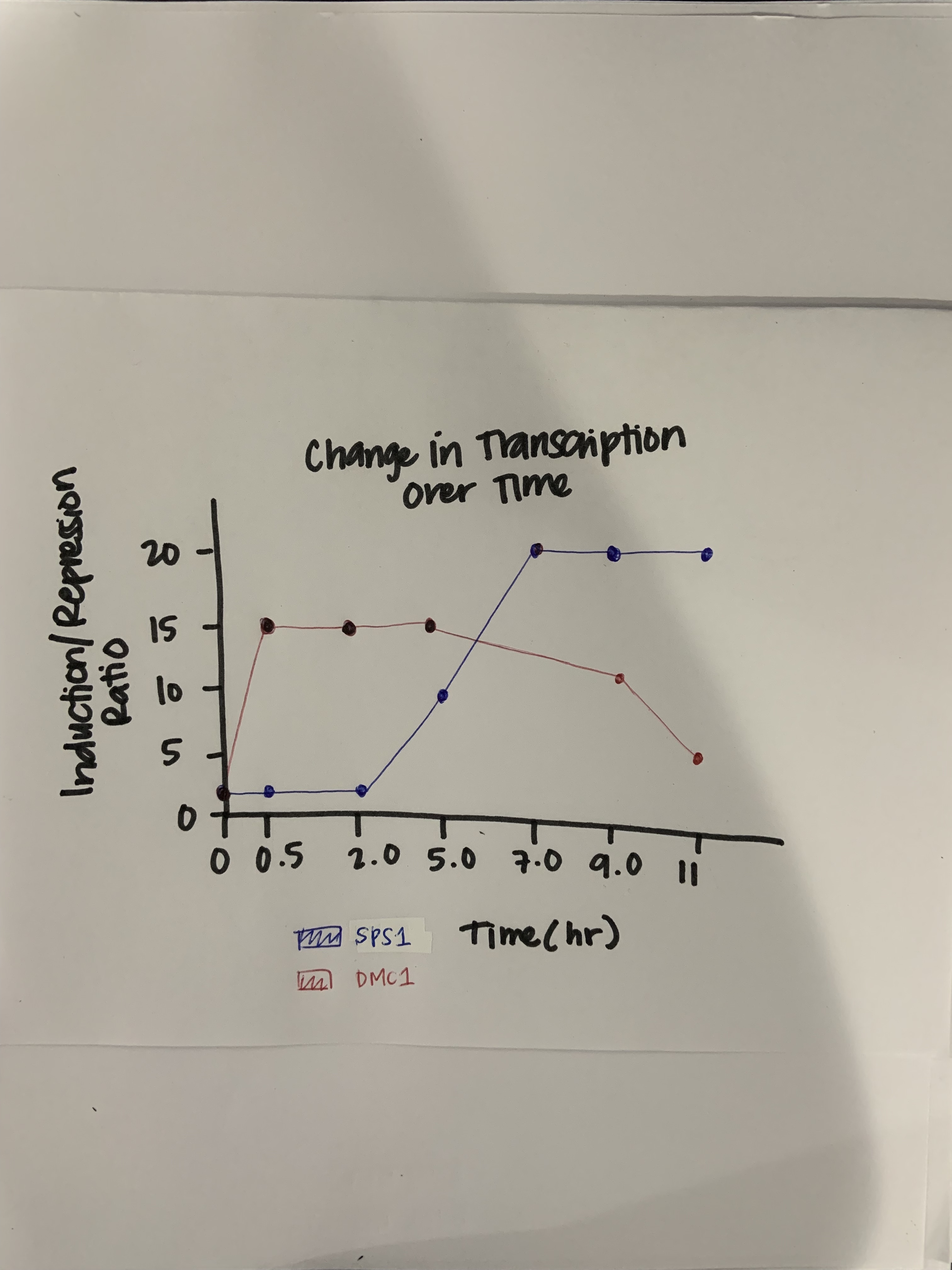
- (Question 5, p. 110) Choose two genes from Figure 4.6b (PDF of figures on Brightspace) and draw a graph to represent the change in transcription over time. You can either create your plot in Excel and put the image up on your wiki page or you can do it by hand and upload a picture or scan.
- (Question 6b, p. 110) Look at Figure 4.7, which depicts the loss of oxygen over time and the transcriptional response of three genes. These data are the ratios of transcription for genes X, Y, and Z during the depletion of oxygen. Using the color scale from Figure 4.6, determine the color for each ratio in Figure 4.7b. (Use the nomenclature "bright green", "medium green", "dim green", "black", "dim red", "medium red", or "bright red" for your answers.)
- Colors:
- Gene X - 1 hour: black - 3 hour: dim red - 5 hour: black - 9 hour: dim green
- Gene Y - 1 hour: black - 3 hour: medium red - 5 hour: black - 9 hour: dim green
- Gene Z - 1 hour: black - 3 hour: dim red - 3 hour: dim red - 9 hour: dim red
- Colors:
- (Question 7, p. 110) Were any of the genes in Figure 4.7b transcribed similarly? If so, which ones were transcribed similarly to which ones?
- Genes X and Y were transcribed most similarly - both black at 1 hour, then red at 3 hour, and then both ratios decreased. The ratios refer to the rate of transcription and depress is due to the lack of oxygen.
- (Question 9, p. 118) Why would most spots be yellow at the first time point? I.e., what is the technical reason that spots show up as yellow - where does the yellow color come from? And, what would be the biological reason that the experiment resulted in most spots being yellow?
- Yellow spots are a way to visualize the green to red ratio (1:1). It comes from storing two color images (green and red) on a computer and merging them to indicate which genes are both expressed in a transcriptome. Most are yellow spots because the it is at its initial time point (t=0) and has not been induced or repressed yet.
- (Question 10, p. 118) Go to the Saccharomyces Genome Database and search for the gene TEF4; you will see it is involved in translation. Look at the time point labeled OD 3.7 in Figure 4.12, and find the TEF4 spot. Over the course of this experiment, was TEF4 induced or repressed? Hypothesize why TEF4’s change in expression was part of the cell’s response to a reduction in available glucose (i.e., the only available food).
- After searching on the genome database, the TEF4 gene turns from yellow towards green which shows that the gene is repressed over the course of the experiment.
- (Question, 11, p. 120) Why would TCA cycle genes be induced if the glucose supply is running out?
- In the TCA cycle, genes are induced even when the glucose supply is running out to change the expression levels of the groups (to increase level of production rather than to convert glucose into stored sugars.
- (Question 12, p. 120) What mechanism could the genome use to ensure genes for enzymes in a common pathway are induced or repressed simultaneously?
- The genome could uncover new pathways in response to the changing conditions of the induced and/or repressed genes, or cluster unknown genes with known genes so they are regulated the same way and function similarly.
- (Question 13, p. 121) Consider a microarray experiment where cells deleted for the repressor TUP1 were subjected to the same experiment of a timecourse of glucose depletion where cells at t0 (plenty of glucose available) are labeled green and cells at later timepoints (glucose depleted) are labeled red. What color would you expect the spots that represented glucose-repressed genes to be in the later time points of this experiment?
- - At a later time point in the experiment, we would expect the spots that represent glucose-repressed genes to be red.
- (Question 14, p. 121) Consider a microarray experiment where cells that overexpress the transcription factor Yap1 were subjected to the same experiment of a timecourse of glucose depletion where cells at t0 (plenty of glucose available) are labeled green and cells at later timepoints (glucose depleted) are labeled red. What color would you expect the spots that represented Yap1 target genes to be in the later time points of this experiment?
- The color that is expected from the spots that represent Yap1p target genes will be red at a later time point in this experiment.
- (Question 16, p. 121) Using the microarray data, how could you verify that you had truly deleted TUP1 or overexpressed YAP1 in the experiments described in questions 8 and 9?
- Based on the analysis of the experiment on the two mutant strains, we could verify the overexpression of the YAP1 gene results in many red spots as answered in questions 8 and 9. Likewise, in the deletion of TUP1, there are a large number of red dots seen.
Acknowledgements
I would like to acknowledge Emma Young (User:Eyoung20), my partner for this week's assignment.
Except for what is noted above, this individual journal entry was completed by me and not copied from another source. Knguye66 (talk) 22:17, 9 October 2019 (PDT) Template:knguye66
References
- https://brightspace.lmu.edu/d2l/le/content/90235/viewContent/800687/View
- https://brightspace.lmu.edu/d2l/le/content/90235/viewContent/800686/View
- Campbell, A.M, & Heeyer, L.J. (2003). "Chapter 4: Basic Research with DNA Microarrays," in Discovering Genomics, Proteomics, and Bloinformatics, Cold Spring Harbor Laboratory Press
- Week_6
- Saccharomyces Genome Database