Difference between revisions of "ILT1/YDR090C Week 3"
(edit bold) |
(forgot to add journal entry) |
||
| (47 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
== "Our Favorite Gene": ILT1/YDR090C == | == "Our Favorite Gene": ILT1/YDR090C == | ||
| − | Gene ILT1 / YDR090C, part of the Saccharomyces Cerevisiae family, is an integral membrane protein (IMP) located on Chromosome IV on the reverse strand that has an unknown molecular function and biological process. It has a molecular weight of 34626.8 Da and an amino acid length of 310. It's primary transcript is YDR090C_mRNA, although much like it's function, information on this gene is unknown/assumed. | + | Gene ILT1 / YDR090C, part of the Saccharomyces Cerevisiae family, is an integral membrane protein (IMP) located on Chromosome IV on the reverse strand that has an unknown molecular function and biological process. It has a molecular weight of 34626.8 Da and an amino acid length of 310. It's primary transcript is YDR090C_mRNA, although much like it's function, information on this gene is unknown and/or assumed. |
== Additional Information == | == Additional Information == | ||
| − | ''' ILT1/YDR090C ''' | + | '''What is the standard name, systematic name, and name description for your gene (from SGD)?''' |
| + | * Standard name: ILT1 | ||
| + | * Systematic name: YDR090C | ||
| + | * Name description for gene: Ionic Liquid Tolerance | ||
| + | '''What is the gene ID (identifier) for your gene in all four databases (SGD, NCBI Gene, Ensembl, UniProt)? Provide hyperlinks to the specific pages for your gene in each of the above databases.''' | ||
| + | * Gene ID: | ||
| + | ** SGD - SGD:S000002497 - ''For more information, visit [https://www.yeastgenome.org/locus/S000002497/ Saccharomyces Genome Database]'' | ||
| + | ** NCBI - Gene ID: 851664 - ''For more information, visit [https://www.ncbi.nlm.nih.gov/gene/851664/ NCBI]'' | ||
| + | ** Ensembl - YDR090C - ''For more information, visit [https://uswest.ensembl.org/Saccharomyces_cerevisiae/Gene/Summary?db=core;g=YDR090C;r=IV:625066-625998;t=YDR090C_mRNA/ Ensembl]'' | ||
| + | ** Uniprot - Q03193 (YD090_YEAST) - ''For more information, visit [https://www.uniprot.org/uniprot/Q03193/ UniProtKB]'' | ||
| + | '''What is the DNA sequence of your gene?''' | ||
| + | ATGATCTCTGAAAAGGCTGCTACCGCTTTAGCTACTATAGCAACAGTTTGCTGGTGTGTCCAGCTAATTCCGCAAAT | ||
| + | AATATATAATTGGAAAAAAAAGGACTGTACGGGGCTGCCACCACTGATGATGTTCCTCTGGGTTGTTTCCGGAATTCCAT | ||
| + | TTGCCATATATTTCTGTGTTAGTAAAGGTAATGTCATTTTACAAGTTCAACCACACCTTTTTATGTTCTTCTGTTCTATA | ||
| + | AGTTTCGTCCAATCGTGTTATTATCCACCAATCAGTATGGCGAGATCCAAAATAGTAATGATTGTAGCTGCTATTATTGC | ||
| + | GGCTGATGTTGGCATGGAAGTTGGCTTTATATTATGGCTGAGGCCCCTTTACGAGAAGGGTGTTAAATGGCCCGATCTGA | ||
| + | TTTTTGGTATTTCTGCATCTGTTTTGTTAGCTGTCGGACTGCTTCCACCTTACTTTGAACTAGCCAAAAGGAAAGGTCGT | ||
| + | GTTATTGGCATCAACTTTGCCTTCTTATTCATCGATTCATTAGGTGCTTGGCTATCTATCATCAGTGTTATCCTAGGTAA | ||
| + | TATGGATATTATGGGTATTATATTATACTCAATAGTTGCTGGAATGGAGTTAGGAATTTTCGCGTCTCATTTCATATGGT | ||
| + | GGTGTAGGTTTAGATTCCTAGCTAAAGGCAATACTTTTGACGAAGAAAGTGGCCAAGCTCAAAAGGAAGAACCCGATGAA | ||
| + | AAAATCGAACAAGATATTAGCAAGAGTGACCGAAATGTTACTAACTATAACCTGGATAACTGTTCAATCCCCGACGATGC | ||
| + | TTCGAGCTTTGCAGACGATTTTAATATATACGATAGTACTGATGGGGGGACGTTATCAAGAGCCCAAACATTGCATGCTG | ||
| + | TCCATGGAGTTGTGGTTAGAACAGATCCTGATCGTTATTCGAGGCTAAGTGTGTAA | ||
| + | '''What is the protein sequence corresponding to your gene?''' | ||
| + | * Frame 1, located in the image, is the protein sequence that corresponds to the gene. | ||
| + | '''What is the function of your gene?''' | ||
| + | * The function of this gene is unknown. Information on this gene is putative. | ||
| + | '''What was different about the information provided about your gene in each of the parent databases?''' | ||
| + | '''Were there differences in content, the information or data itself?''' | ||
| + | *There weren't many big differences in the content between each of the databases. For instance, NCBI showed assumed gene sequence. Also, there was more information displayed in NCBI compare to anything else. Since this is a gene that is missing a lot of information due to lack of research, most databases provide about the exact same information. Each one consists of the SGD ID and lineage. The function type from each database is the same. There aren't huge differences that can be displayed. | ||
| + | '''Were there differences in presentation of the information?''' | ||
| + | * The differences in presentation was the different color schemes between the databases. For example, UniProt has this very bright blue color, whereas NCBI is this gray color. SGD seems to be the only database that tends to be very straightforward on the information. It is to the point and big enough for specific information. UniProt is another database that can be used easily and has a great representation. | ||
| + | '''Why did you choose your particular gene? i.e., why is it interesting to you and your partner?''' | ||
| + | * This particular gene was chosen because much of its information has yet to be discovered. It is a protein with unknown function and very few research has been conducted and found on this gene. This sparked interest to us because it highlighted the very point of science and why we are biology majors: to discover (using research and evidence) to further identify and/or understand a distinct mechanism, organism, and/or gene. | ||
| − | + | == Images == | |
| − | + | '''Include an image related to your gene (be careful that you do not violate any copyright restrictions!)''' | |
| − | + | *Take a screenshot of your results, display it on your wiki page, and state which frame it is. | |
| − | + | * Please make the image something scientific (not like the random images seen on the SGD blog posts). | |
| − | + | * If a 3D structure of the protein your gene encodes is available, you can choose to embed a rotating image of the structure on your page using the FirstGlance in Jmol software. This is optional, a different static image would be OK, too. | |
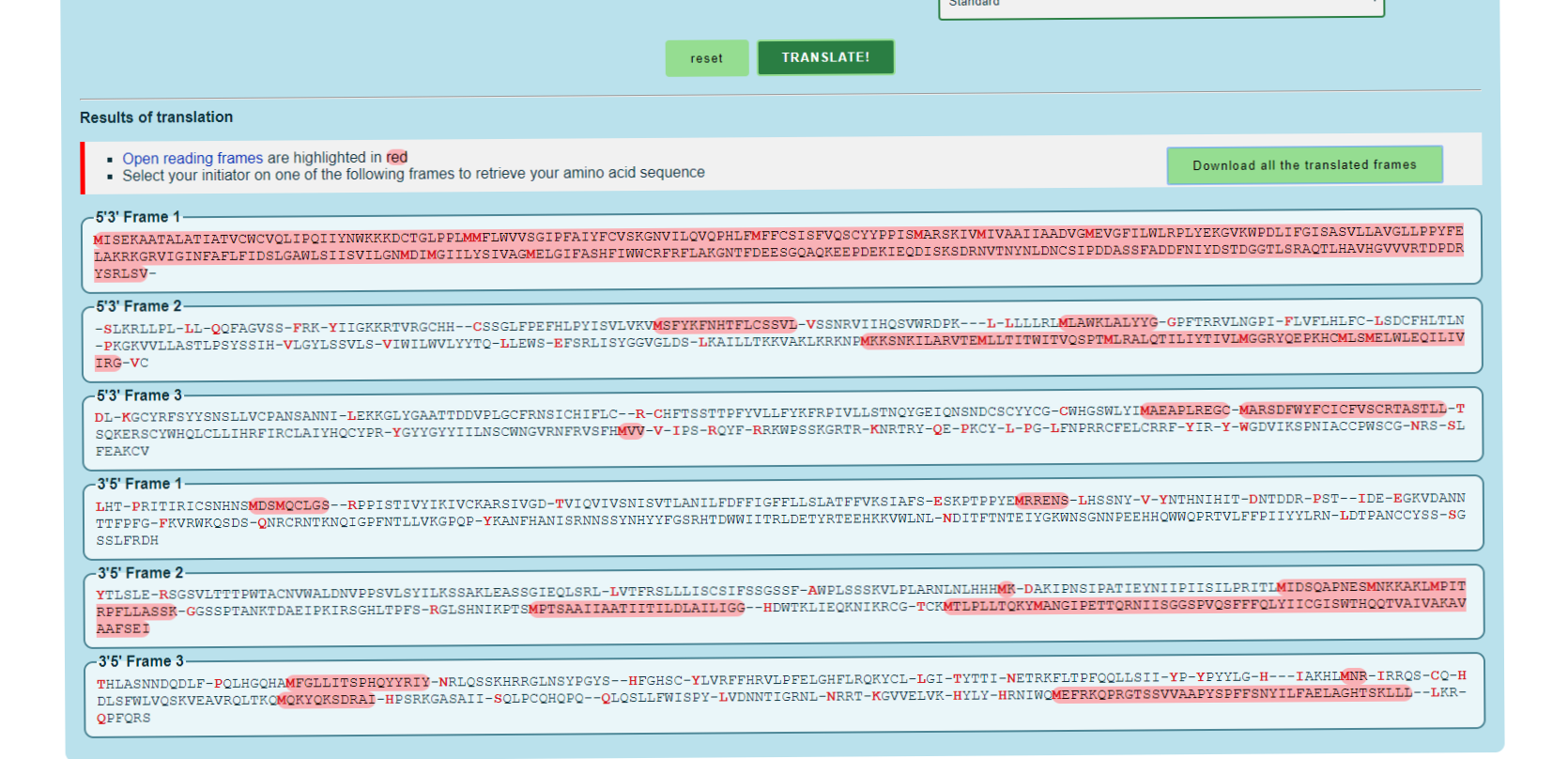
| − | + | [[Image:YDR090CProtein.png|thumb|center|600px|Results containing protein sequence for YDR090C. Translated from ExPASy.]] | |
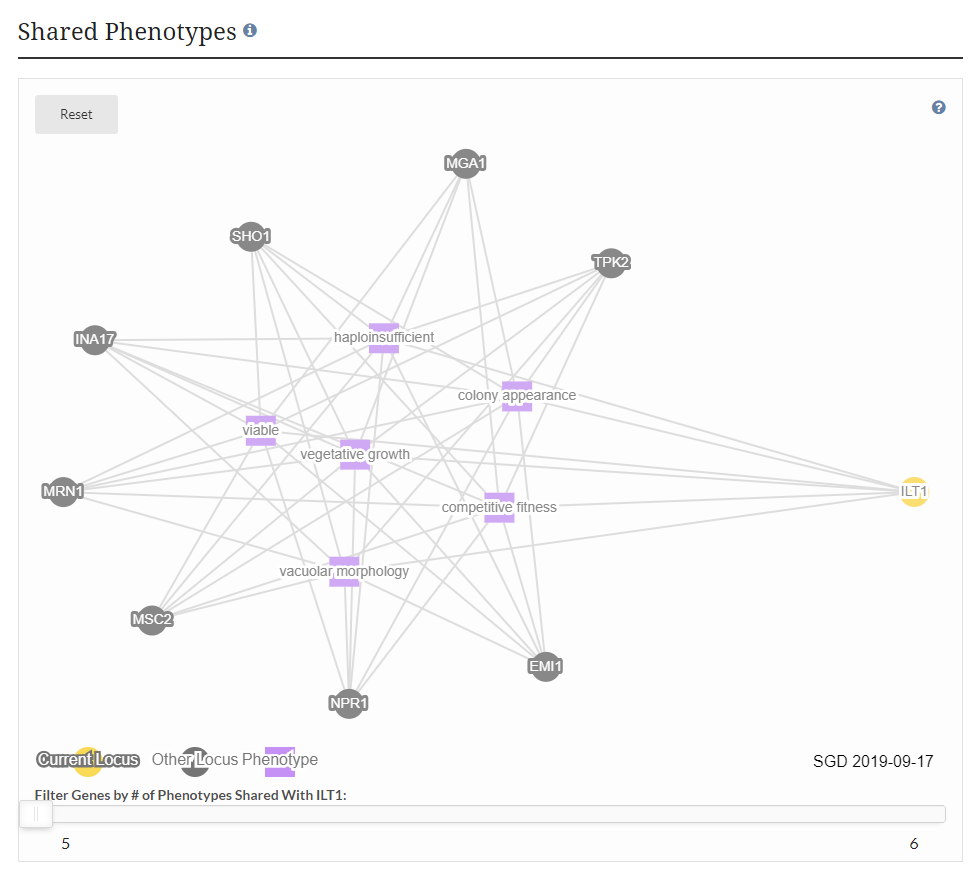
| − | + | [[File:Shared Phenotypes.png|600px|thumb|center|This diagram displays phenotype observables (purple squares) that are shared between the given gene (yellow circle) and other genes (gray circles) based on the number of phenotype observables shared (adjustable using the slider at the bottom).]] | |
| − | |||
| − | |||
| − | |||
| − | ''' | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
== Acknowledgements == | == Acknowledgements == | ||
* This section is in acknowledgement to partners, Kaitlyn Nguyen [[User:Knguye66]] and Iliana Crespin [[User:Icrespin]]. | * This section is in acknowledgement to partners, Kaitlyn Nguyen [[User:Knguye66]] and Iliana Crespin [[User:Icrespin]]. | ||
| − | + | * Kaitlyn did a lot of work in this wiki page from organizing to summarizing the gene. Iliana helped with organization and researching what this gene is about. | |
| − | Except for what is noted above, this individual journal entry was completed by me and not copied from another source. | + | *"Except for what is noted above, this individual journal entry was completed by me and not copied from another source." [[User:Icrespin|Icrespin]] ([[User talk:Icrespin|talk]]) 15:32, 17 September 2019 (PDT) [[User:Knguye66|Knguye66]] ([[User talk:Knguye66|talk]]) 15:47, 17 September 2019 (PDT) |
== References == | == References == | ||
| + | * Dunn, B., & MacPherson, K. (2018, September 24). New & Noteworthy. Retrieved from https://www.yeastgenome.org/blog/category/research-spotlight | ||
| + | * Gene: YDR090C. (n.d.). Retrieved from https://uswest.ensembl.org/Saccharomyces_cerevisiae/Gene/Summary?db=core;g=YDR090C;r=IV:625066-625998;t=YDR090C_mRNA | ||
* ILT1. (n.d.). Retrieved from https://www.yeastgenome.org/locus/S000002497 | * ILT1. (n.d.). Retrieved from https://www.yeastgenome.org/locus/S000002497 | ||
| + | * ILT1. (n.d.). Retrieved from https://www.yeastgenome.org/locus/S000002497/phenotype | ||
| + | * SIB Swiss Institute of Bioinformatics. (n.d.). SIB Swiss Institute of Bioinformatics. Retrieved from https://web.expasy.org/translate/ | ||
| + | * UniProt ConsortiumEuropean Bioinformatics InstituteProtein Information ResourceSIB Swiss Institute of Bioinformatics. (2019, May 8). Uncharacterized membrane protein YDR090C. Retrieved from https://www.uniprot.org/uniprot/Q03193 | ||
| + | *Week 3 Assignment page is: LMU BioDB 2019. (2019). Week 3. Retrieved September 17, 2019, from https://xmlpipedb.cs.lmu.edu/biodb/fall2019/index.php/Week_3 | ||
* YDR090C hypothetical protein [Saccharomyces cerevisiae S288C] - Gene - NCBI. (n.d.). Retrieved from https://www.ncbi.nlm.nih.gov/gene/851664 | * YDR090C hypothetical protein [Saccharomyces cerevisiae S288C] - Gene - NCBI. (n.d.). Retrieved from https://www.ncbi.nlm.nih.gov/gene/851664 | ||
| − | |||
| − | |||
''' Wikipedia Techniques: ''' | ''' Wikipedia Techniques: ''' | ||
* [Cedrick May]. (2014, August 26). Adding Hyperlinks to a Wiki Page [Video file]. Retrieved from https://www.youtube.com/watch?v=J50espeRSs8&t=137s | * [Cedrick May]. (2014, August 26). Adding Hyperlinks to a Wiki Page [Video file]. Retrieved from https://www.youtube.com/watch?v=J50espeRSs8&t=137s | ||
| + | * Extended image syntax. (2019, July 6). Retrieved from https://en.wikipedia.org/wiki/Wikipedia:Extended_image_syntax | ||
* Hyperlink. (2019, September 8). Retrieved from https://en.wikipedia.org/wiki/Hyperlink | * Hyperlink. (2019, September 8). Retrieved from https://en.wikipedia.org/wiki/Hyperlink | ||
| + | |||
| + | [[Category:Journal Entry]] | ||
Latest revision as of 19:59, 9 October 2019
Contents
"Our Favorite Gene": ILT1/YDR090C
Gene ILT1 / YDR090C, part of the Saccharomyces Cerevisiae family, is an integral membrane protein (IMP) located on Chromosome IV on the reverse strand that has an unknown molecular function and biological process. It has a molecular weight of 34626.8 Da and an amino acid length of 310. It's primary transcript is YDR090C_mRNA, although much like it's function, information on this gene is unknown and/or assumed.
Additional Information
What is the standard name, systematic name, and name description for your gene (from SGD)?
- Standard name: ILT1
- Systematic name: YDR090C
- Name description for gene: Ionic Liquid Tolerance
What is the gene ID (identifier) for your gene in all four databases (SGD, NCBI Gene, Ensembl, UniProt)? Provide hyperlinks to the specific pages for your gene in each of the above databases.
- Gene ID:
- SGD - SGD:S000002497 - For more information, visit Saccharomyces Genome Database
- NCBI - Gene ID: 851664 - For more information, visit NCBI
- Ensembl - YDR090C - For more information, visit Ensembl
- Uniprot - Q03193 (YD090_YEAST) - For more information, visit UniProtKB
What is the DNA sequence of your gene?
ATGATCTCTGAAAAGGCTGCTACCGCTTTAGCTACTATAGCAACAGTTTGCTGGTGTGTCCAGCTAATTCCGCAAAT AATATATAATTGGAAAAAAAAGGACTGTACGGGGCTGCCACCACTGATGATGTTCCTCTGGGTTGTTTCCGGAATTCCAT TTGCCATATATTTCTGTGTTAGTAAAGGTAATGTCATTTTACAAGTTCAACCACACCTTTTTATGTTCTTCTGTTCTATA AGTTTCGTCCAATCGTGTTATTATCCACCAATCAGTATGGCGAGATCCAAAATAGTAATGATTGTAGCTGCTATTATTGC GGCTGATGTTGGCATGGAAGTTGGCTTTATATTATGGCTGAGGCCCCTTTACGAGAAGGGTGTTAAATGGCCCGATCTGA TTTTTGGTATTTCTGCATCTGTTTTGTTAGCTGTCGGACTGCTTCCACCTTACTTTGAACTAGCCAAAAGGAAAGGTCGT GTTATTGGCATCAACTTTGCCTTCTTATTCATCGATTCATTAGGTGCTTGGCTATCTATCATCAGTGTTATCCTAGGTAA TATGGATATTATGGGTATTATATTATACTCAATAGTTGCTGGAATGGAGTTAGGAATTTTCGCGTCTCATTTCATATGGT GGTGTAGGTTTAGATTCCTAGCTAAAGGCAATACTTTTGACGAAGAAAGTGGCCAAGCTCAAAAGGAAGAACCCGATGAA AAAATCGAACAAGATATTAGCAAGAGTGACCGAAATGTTACTAACTATAACCTGGATAACTGTTCAATCCCCGACGATGC TTCGAGCTTTGCAGACGATTTTAATATATACGATAGTACTGATGGGGGGACGTTATCAAGAGCCCAAACATTGCATGCTG TCCATGGAGTTGTGGTTAGAACAGATCCTGATCGTTATTCGAGGCTAAGTGTGTAA
What is the protein sequence corresponding to your gene?
- Frame 1, located in the image, is the protein sequence that corresponds to the gene.
What is the function of your gene?
- The function of this gene is unknown. Information on this gene is putative.
What was different about the information provided about your gene in each of the parent databases? Were there differences in content, the information or data itself?
- There weren't many big differences in the content between each of the databases. For instance, NCBI showed assumed gene sequence. Also, there was more information displayed in NCBI compare to anything else. Since this is a gene that is missing a lot of information due to lack of research, most databases provide about the exact same information. Each one consists of the SGD ID and lineage. The function type from each database is the same. There aren't huge differences that can be displayed.
Were there differences in presentation of the information?
- The differences in presentation was the different color schemes between the databases. For example, UniProt has this very bright blue color, whereas NCBI is this gray color. SGD seems to be the only database that tends to be very straightforward on the information. It is to the point and big enough for specific information. UniProt is another database that can be used easily and has a great representation.
Why did you choose your particular gene? i.e., why is it interesting to you and your partner?
- This particular gene was chosen because much of its information has yet to be discovered. It is a protein with unknown function and very few research has been conducted and found on this gene. This sparked interest to us because it highlighted the very point of science and why we are biology majors: to discover (using research and evidence) to further identify and/or understand a distinct mechanism, organism, and/or gene.
Images
Include an image related to your gene (be careful that you do not violate any copyright restrictions!)
- Take a screenshot of your results, display it on your wiki page, and state which frame it is.
- Please make the image something scientific (not like the random images seen on the SGD blog posts).
- If a 3D structure of the protein your gene encodes is available, you can choose to embed a rotating image of the structure on your page using the FirstGlance in Jmol software. This is optional, a different static image would be OK, too.
Acknowledgements
- This section is in acknowledgement to partners, Kaitlyn Nguyen User:Knguye66 and Iliana Crespin User:Icrespin.
- Kaitlyn did a lot of work in this wiki page from organizing to summarizing the gene. Iliana helped with organization and researching what this gene is about.
- "Except for what is noted above, this individual journal entry was completed by me and not copied from another source." Icrespin (talk) 15:32, 17 September 2019 (PDT) Knguye66 (talk) 15:47, 17 September 2019 (PDT)
References
- Dunn, B., & MacPherson, K. (2018, September 24). New & Noteworthy. Retrieved from https://www.yeastgenome.org/blog/category/research-spotlight
- Gene: YDR090C. (n.d.). Retrieved from https://uswest.ensembl.org/Saccharomyces_cerevisiae/Gene/Summary?db=core;g=YDR090C;r=IV:625066-625998;t=YDR090C_mRNA
- ILT1. (n.d.). Retrieved from https://www.yeastgenome.org/locus/S000002497
- ILT1. (n.d.). Retrieved from https://www.yeastgenome.org/locus/S000002497/phenotype
- SIB Swiss Institute of Bioinformatics. (n.d.). SIB Swiss Institute of Bioinformatics. Retrieved from https://web.expasy.org/translate/
- UniProt ConsortiumEuropean Bioinformatics InstituteProtein Information ResourceSIB Swiss Institute of Bioinformatics. (2019, May 8). Uncharacterized membrane protein YDR090C. Retrieved from https://www.uniprot.org/uniprot/Q03193
- Week 3 Assignment page is: LMU BioDB 2019. (2019). Week 3. Retrieved September 17, 2019, from https://xmlpipedb.cs.lmu.edu/biodb/fall2019/index.php/Week_3
- YDR090C hypothetical protein [Saccharomyces cerevisiae S288C] - Gene - NCBI. (n.d.). Retrieved from https://www.ncbi.nlm.nih.gov/gene/851664
Wikipedia Techniques:
- [Cedrick May]. (2014, August 26). Adding Hyperlinks to a Wiki Page [Video file]. Retrieved from https://www.youtube.com/watch?v=J50espeRSs8&t=137s
- Extended image syntax. (2019, July 6). Retrieved from https://en.wikipedia.org/wiki/Wikipedia:Extended_image_syntax
- Hyperlink. (2019, September 8). Retrieved from https://en.wikipedia.org/wiki/Hyperlink