Difference between revisions of "Individual Journal Page"
Jump to navigation
Jump to search
(linked journal, wrote acknowledgments) |
(→References: edited references) |
||
| (43 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | ==Week 1== | + | ==Imacarae Week 1== |
===Shared Journal Assignment=== | ===Shared Journal Assignment=== | ||
*[[https://xmlpipedb.cs.lmu.edu/biodb/fall2019/index.php/Class_Journal_Week_1 Week 1 Journal]] | *[[https://xmlpipedb.cs.lmu.edu/biodb/fall2019/index.php/Class_Journal_Week_1 Week 1 Journal]] | ||
===Acknowledgments=== | ===Acknowledgments=== | ||
| − | *I worked with my partner, [[https://xmlpipedb.cs.lmu.edu/biodb/fall2019/index.php/User:Cdomin12 Christina Dominguez]], in class. We helped each other with the format of editing User and Template pages, as well as helped clarify shared ambiguities of the Week 1 assignment. There was no other contact outside of class or through phone. | + | *I worked with my homework partner, [[https://xmlpipedb.cs.lmu.edu/biodb/fall2019/index.php/User:Cdomin12 Christina Dominguez]], in class. We helped each other with the format of editing User and Template pages, as well as helped clarify shared ambiguities of the Week 1 assignment. There was no other contact outside of class or through phone. |
| − | *I | + | *[[https://xmlpipedb.cs.lmu.edu/biodb/fall2019/index.php/User:Dmadere DeLisa Madere]] and I consulted with each other on formatting of pictures and links to pages. |
| + | *Except what is noted above, this individual journal entry was completed by me and not copied from another source. | ||
| + | [[User:Imacarae|Imacarae]] ([[User talk:Imacarae|talk]]) 18:31, 4 September 2019 (PDT) | ||
| + | |||
===References=== | ===References=== | ||
| − | * | + | *Instructions for Wiki layout and assignment provided by LMU BioDB 2019. (2019). Week 1. Retrieved August 29, 2017, from [[https://xmlpipedb.cs.lmu.edu/biodb/fall2019/index.php/Week_1 https://xmlpipedb.cs.lmu.edu/biodb/fall2019/index.php/Week_1]] |
| + | |||
| + | ==Imacarae Week 2== | ||
| + | |||
| + | ===Biochemistry:=== | ||
| + | ====Purpose==== | ||
| + | *To understand how sequences of amino acids affect expression of colors in flowers. | ||
| + | *To use mechanics of genetics, biochemistry, molecular biology, and evolution in studying this expression. | ||
| + | |||
| + | ====Methods and Results==== | ||
| + | #Downloaded and opened [[http://aipotu.umb.edu/ Aipotu]]. | ||
| + | #Opened "Biochemistry" tab to view amino acid sequences of Green-1 and White and compare them. | ||
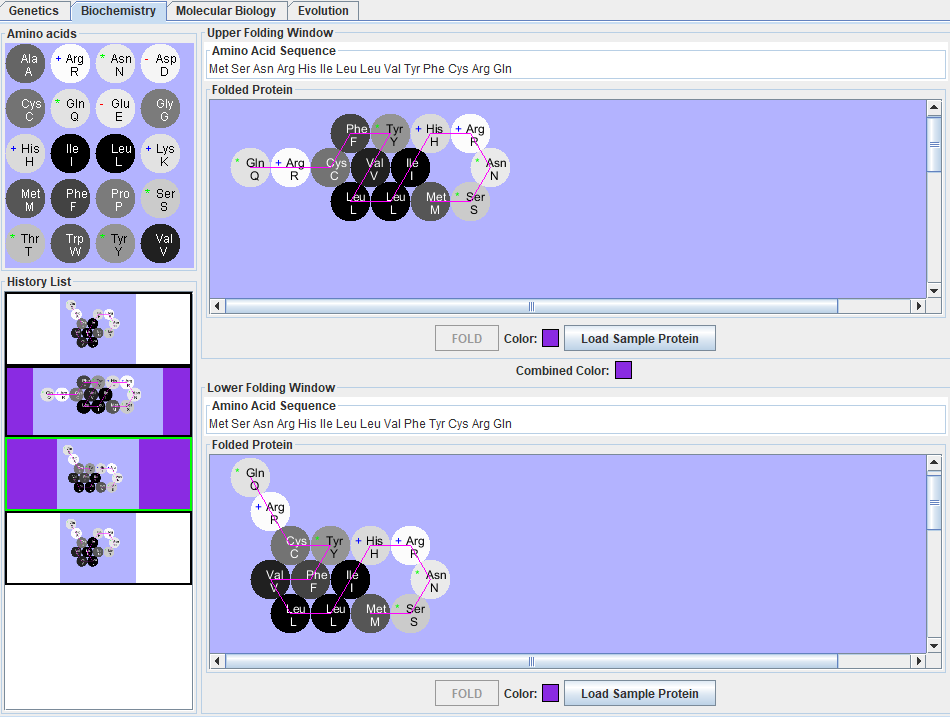
| + | #*Results to ST(a): For alleles green and white, there are two differences in amino acids. They are a change from Tyrosine to Valine and from Tryptophan to Cysteine in Green-1 to White respectively. | ||
| + | #*Results to ST(b,c,&d): What makes the protein pigmented are the change in Val in the 10th position to a different amino acid.What makes the protein a particular color is the change in differences in the 10th or 11th amino acid in the sequence. Colors combine by adding the amino acid that made them different from another color into the original sequence. | ||
| + | #Started on the "Genetics" tab to cross organisms to produce "Purple" organism. | ||
| + | #*Crossed organism Green-2 to Red. | ||
| + | #*Selected organism 2-8 as "Purple" organism. | ||
| + | #Inserted amino acid Tyr between amino acids Phe and Cys to create purple protein. | ||
| + | #Inserted amino acid Phe between amino acids Tyr and Cyst to create additional protein. | ||
| + | #*Results to ST(f): The purple protein constructed is sequenced as MSNRHILLVYFCRQ. | ||
| + | #Pure-color, homozygous proteins could be created by the use of amino acids Phe, Tyr, and Trp. | ||
| + | #*These amino acids contain aromatic rings which allows for pigment. | ||
| + | |||
| + | ====Data and Files==== | ||
| + | [[File:ImacaraePurpleprotein.jpg.png|350px|right]] | ||
| + | |||
| + | {| class="wikitable" | ||
| + | |+ Characteristics of the Four Starting Organisms | ||
| + | ! Organism | ||
| + | ! Allele | ||
| + | ! Color | ||
| + | ! Amino Acid Sequence | ||
| + | |- | ||
| + | ! scope=row | Green-1 | ||
| + | | CgCg | ||
| + | | Green | ||
| + | | MSNRHILLVYWRQ | ||
| + | |- | ||
| + | ! scope=row | Green-2 | ||
| + | | CbCy | ||
| + | | Green | ||
| + | | MSNRHILLVYCRQ | ||
| + | |- | ||
| + | ! scope=row | Red | ||
| + | | CrCr | ||
| + | | Red | ||
| + | | MSNRHILLVFCRQ | ||
| + | |- | ||
| + | ! scope=row | White | ||
| + | | CwCw | ||
| + | | White | ||
| + | | MSNRHILLVVCRQ | ||
| + | |} | ||
| + | |||
| + | ====Scientific Conclusion==== | ||
| + | *The main finding of this project was that not only can an organism's alleles be determined by "trial and error" breeding, but its protein that codes for color can be directly changed. | ||
| + | **The addition/deletion of specific amino acids will allow for the creation of pure-color proteins. | ||
| + | *Yes, this fulfilled the purpose of the project. Because of this, I better understand how protein sequence, and biochemistry as a whole, is directly correlated with phenotypic expression. | ||
| + | |||
| + | ===Shared Journal Assignment=== | ||
| + | [[https://xmlpipedb.cs.lmu.edu/biodb/fall2019/index.php/Class_Journal_Week_2 Week 2 Journal]] | ||
| + | |||
| + | ===Acknowledgements=== | ||
| + | *I worked with my homework partner, [[https://xmlpipedb.cs.lmu.edu/biodb/fall2019/index.php/User:Dramir36 David Ramirez]], in class. We worked on learning Aipotu, and how it worked, as well answering the questions in the Biochemistry assignment. We met outside of class on two separate occasions to help each other with figuring out how to add amino acids and get results. | ||
| + | *One of our Biochemistry partners, [[https://xmlpipedb.cs.lmu.edu/biodb/fall2019/index.php/User:Marmas Michael Armas]], did help us with creating pure-color proteins by providing us information on aromatic amino acids that give rise to pigment. | ||
| + | *Except with what is noted above, this individual journal entry was completed by me and not copied from another source. | ||
| + | [[User:Imacarae|Imacarae]] ([[User talk:Imacarae|talk]]) 23:20, 10 September 2019 (PDT) | ||
| + | |||
| + | ===References=== | ||
| + | *Program used: Aipotu. (2017). Retrieved September 10, 2019, from [[http://aipotu.umb.edu/ http://aipotu.umb.edu/]]. | ||
| + | *Table/image formatting provided by WikiHelp. (2019). Retrieved September 10, 2019, from [[https://en.wikipedia.org/wiki/Wikipedia:Extended_image_syntax https://en.wikipedia.org/wiki/Wikipedia:Extended_image_syntax]] and [[https://en.wikipedia.org/wiki/Help:Table https://en.wikipedia.org/wiki/Help:Table]]. | ||
| + | *Instructions for Week 2 assignment provided by LMU BioDB 2019. (2019). Week 2. Retrieved September 10, 2019, from [[https://xmlpipedb.cs.lmu.edu/biodb/fall2019/index.php/Week_2 https://xmlpipedb.cs.lmu.edu/biodb/fall2019/index.php/Week_2]]. | ||
| + | |||
| + | ==Week 3== | ||
| + | |||
| + | ==Week 4== | ||
| + | |||
| + | ==Week 5== | ||
| + | |||
| + | ==Week 6== | ||
| + | |||
| + | ==Week 7== | ||
| + | |||
| + | ==Week 8== | ||
Latest revision as of 22:33, 10 September 2019
Imacarae Week 1
Acknowledgments
- I worked with my homework partner, [Christina Dominguez], in class. We helped each other with the format of editing User and Template pages, as well as helped clarify shared ambiguities of the Week 1 assignment. There was no other contact outside of class or through phone.
- [DeLisa Madere] and I consulted with each other on formatting of pictures and links to pages.
- Except what is noted above, this individual journal entry was completed by me and not copied from another source.
Imacarae (talk) 18:31, 4 September 2019 (PDT)
References
- Instructions for Wiki layout and assignment provided by LMU BioDB 2019. (2019). Week 1. Retrieved August 29, 2017, from [https://xmlpipedb.cs.lmu.edu/biodb/fall2019/index.php/Week_1]
Imacarae Week 2
Biochemistry:
Purpose
- To understand how sequences of amino acids affect expression of colors in flowers.
- To use mechanics of genetics, biochemistry, molecular biology, and evolution in studying this expression.
Methods and Results
- Downloaded and opened [Aipotu].
- Opened "Biochemistry" tab to view amino acid sequences of Green-1 and White and compare them.
- Results to ST(a): For alleles green and white, there are two differences in amino acids. They are a change from Tyrosine to Valine and from Tryptophan to Cysteine in Green-1 to White respectively.
- Results to ST(b,c,&d): What makes the protein pigmented are the change in Val in the 10th position to a different amino acid.What makes the protein a particular color is the change in differences in the 10th or 11th amino acid in the sequence. Colors combine by adding the amino acid that made them different from another color into the original sequence.
- Started on the "Genetics" tab to cross organisms to produce "Purple" organism.
- Crossed organism Green-2 to Red.
- Selected organism 2-8 as "Purple" organism.
- Inserted amino acid Tyr between amino acids Phe and Cys to create purple protein.
- Inserted amino acid Phe between amino acids Tyr and Cyst to create additional protein.
- Results to ST(f): The purple protein constructed is sequenced as MSNRHILLVYFCRQ.
- Pure-color, homozygous proteins could be created by the use of amino acids Phe, Tyr, and Trp.
- These amino acids contain aromatic rings which allows for pigment.
Data and Files
| Organism | Allele | Color | Amino Acid Sequence |
|---|---|---|---|
| Green-1 | CgCg | Green | MSNRHILLVYWRQ |
| Green-2 | CbCy | Green | MSNRHILLVYCRQ |
| Red | CrCr | Red | MSNRHILLVFCRQ |
| White | CwCw | White | MSNRHILLVVCRQ |
Scientific Conclusion
- The main finding of this project was that not only can an organism's alleles be determined by "trial and error" breeding, but its protein that codes for color can be directly changed.
- The addition/deletion of specific amino acids will allow for the creation of pure-color proteins.
- Yes, this fulfilled the purpose of the project. Because of this, I better understand how protein sequence, and biochemistry as a whole, is directly correlated with phenotypic expression.
Acknowledgements
- I worked with my homework partner, [David Ramirez], in class. We worked on learning Aipotu, and how it worked, as well answering the questions in the Biochemistry assignment. We met outside of class on two separate occasions to help each other with figuring out how to add amino acids and get results.
- One of our Biochemistry partners, [Michael Armas], did help us with creating pure-color proteins by providing us information on aromatic amino acids that give rise to pigment.
- Except with what is noted above, this individual journal entry was completed by me and not copied from another source.
Imacarae (talk) 23:20, 10 September 2019 (PDT)
References
- Program used: Aipotu. (2017). Retrieved September 10, 2019, from [http://aipotu.umb.edu/].
- Table/image formatting provided by WikiHelp. (2019). Retrieved September 10, 2019, from [https://en.wikipedia.org/wiki/Wikipedia:Extended_image_syntax] and [https://en.wikipedia.org/wiki/Help:Table].
- Instructions for Week 2 assignment provided by LMU BioDB 2019. (2019). Week 2. Retrieved September 10, 2019, from [https://xmlpipedb.cs.lmu.edu/biodb/fall2019/index.php/Week_2].