Difference between revisions of "Sulfiknights Deliverables"
(→Statement of Work: uploaded statement of work) |
(→Statement of Work: changed numbering) |
||
| Line 138: | Line 138: | ||
#'''Role as Designer''' | #'''Role as Designer''' | ||
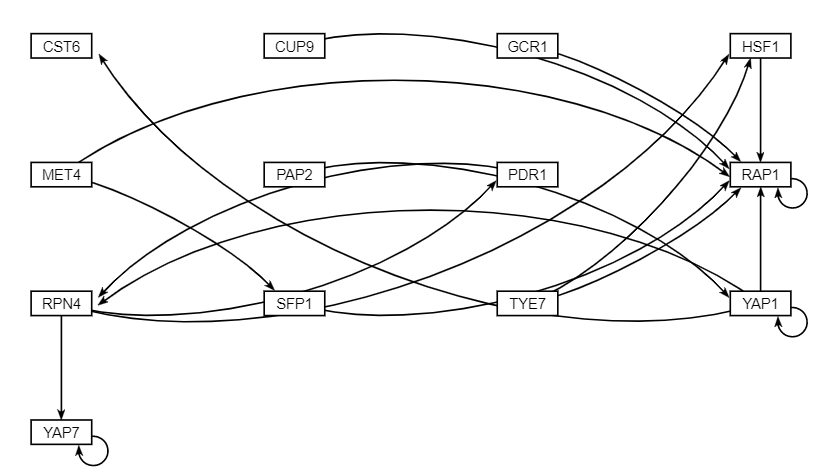
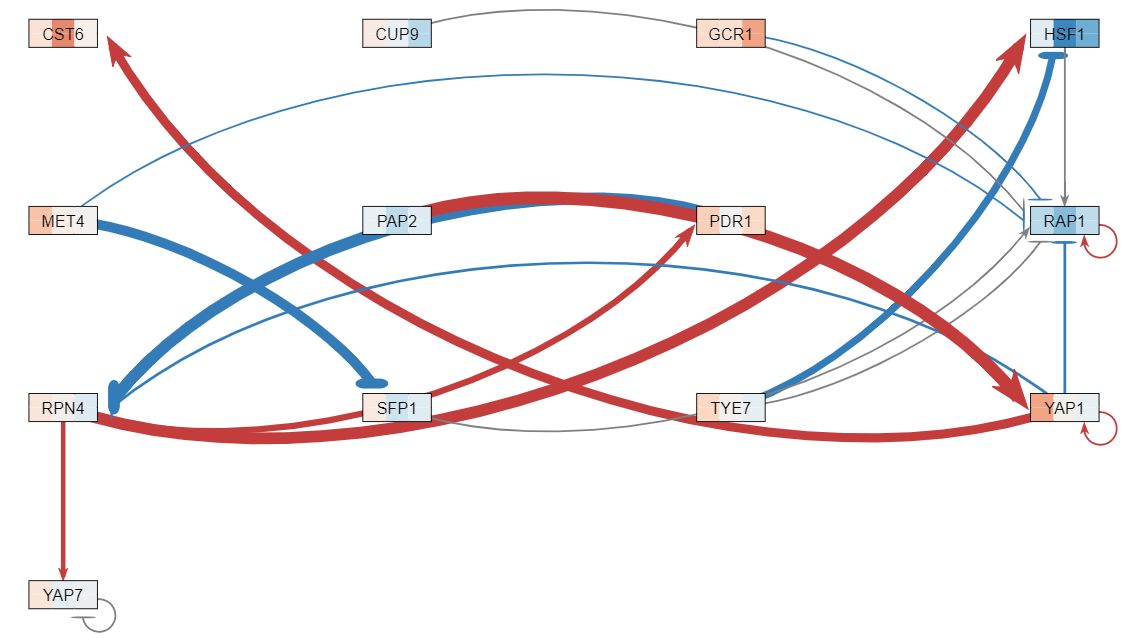
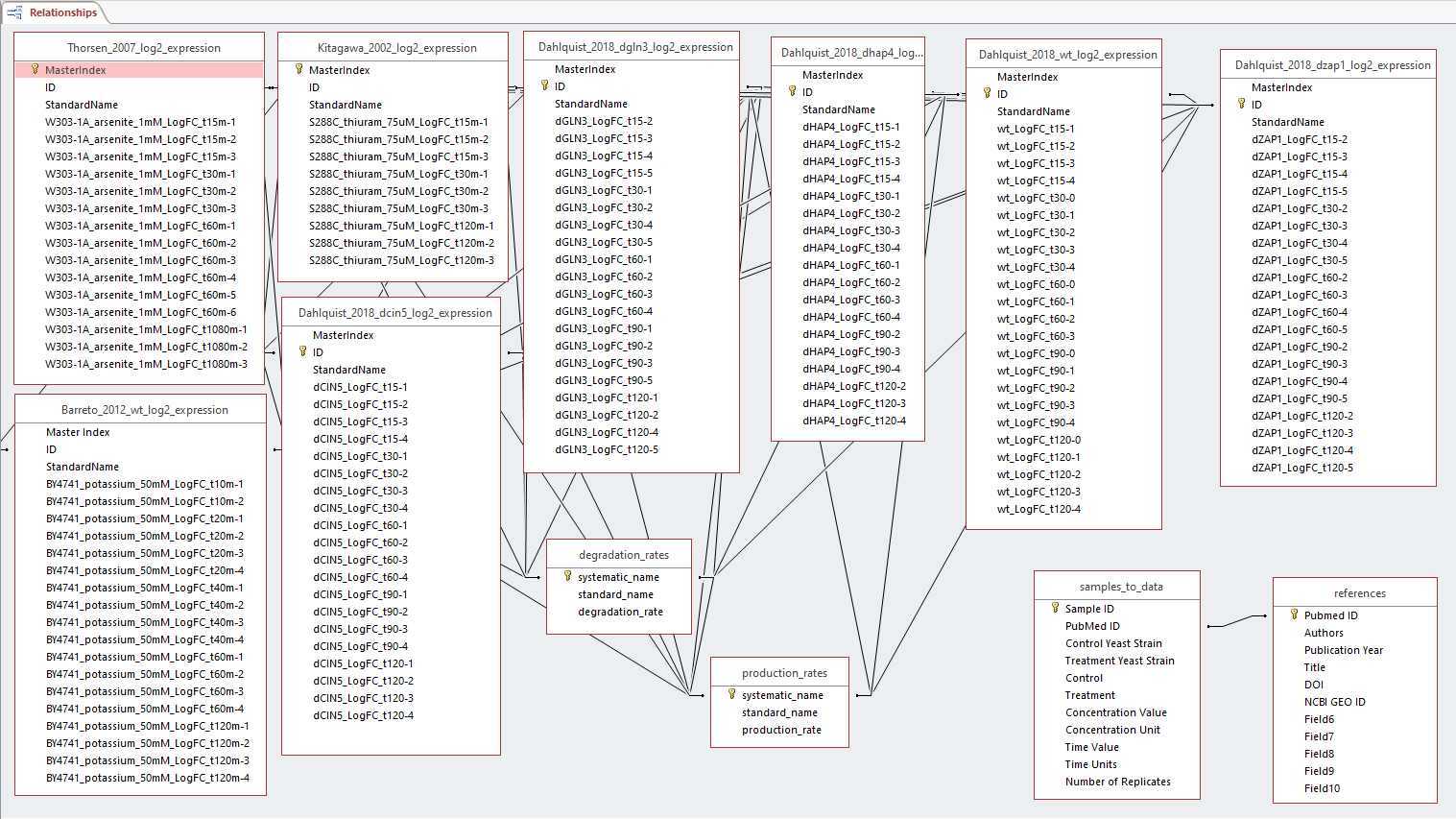
#*My role in this project was a Designer. As a designer, I collaborated with the other coders to come up with a metadata sheet incorporating information regarding the citations of the paper as well as the sample-to-data sheet including the different treatments, yeast strains, timepoints, concentrations, and replicates. After creating the metadata sheet, I worked with the QAs to create the database compiling the datasets from our microarray data for the Data Analysts to analyze further. Once all the data was analyzed, I then worked with the QAs and the other coders to create a database compiling our data, the degradation rates, production rates, metadata table, as well as the other datasets from the other groups (FunGals, Skinny Genes, Dahlquist Lab) to visualize the relationships in a database schema. Each dataset was standardized under common headers such as author_publicationyear_log2_expression, and each dataset had common headers as well representing each replicate such as yeaststrain_treatment_concentration_LogFC_time-replicate. The final database is linked together through a standardized ID that each of the datasets contained. | #*My role in this project was a Designer. As a designer, I collaborated with the other coders to come up with a metadata sheet incorporating information regarding the citations of the paper as well as the sample-to-data sheet including the different treatments, yeast strains, timepoints, concentrations, and replicates. After creating the metadata sheet, I worked with the QAs to create the database compiling the datasets from our microarray data for the Data Analysts to analyze further. Once all the data was analyzed, I then worked with the QAs and the other coders to create a database compiling our data, the degradation rates, production rates, metadata table, as well as the other datasets from the other groups (FunGals, Skinny Genes, Dahlquist Lab) to visualize the relationships in a database schema. Each dataset was standardized under common headers such as author_publicationyear_log2_expression, and each dataset had common headers as well representing each replicate such as yeaststrain_treatment_concentration_LogFC_time-replicate. The final database is linked together through a standardized ID that each of the datasets contained. | ||
| − | |||
#'''Links to Artifacts''' | #'''Links to Artifacts''' | ||
Revision as of 16:51, 12 December 2019
| Sulfiknight Links | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| BIOL Databases Main Page | Sulfiknights: Project Overview Page | Final Project Deliverables Requirements | Sulfiknights: Final Project Deliverables | Members | Project Manager & Quality Assurance: Naomi Tesfaiohannes | Quality Assurance: Joey Nimmers-Minor | Data Analysis: Ivy-Quynh Macaraeg & Marcus Avila | Designer: DeLisa Madere | |
| Assignment Pages | Week 11 | Week 12/13 | Week 15 | ||||||
Contents
- 1 Checklist:
- 2 Group Report
- 3 Individual Statements of Work, Assessments, Reflections
- 4 Group Powerpoint Presentation
- 5 Excel Spreadsheet with ANOVA Results/STEM Formatting
- 6 PowerPoint of ANOVA Table...
- 7 Gene List and GO List files
- 8 YEASTRACT "Rank by TF" Results
- 9 GRNmap input workbook
- 10 GRNmap output workbook and output plots
- 11 MS Access database
- 12 ReadMe
- 13 Query design for populating a GRNmap input workbook from the database
- 14 Electronic notebook corresponding to the microarray results files
- 15 Merge completed databases into a single database
- 16 Final Reflection
Checklist:
- Organized Team deliverables wiki page (or other media (CD or flash drive) with table of contents) INCOMPLETE
- Group Report (.doc, .docx or .pdf file) INCOMPLETE
- Individual statements of work, assessments, reflections (wiki page, .doc, .docx, .pdf, or e-mailed to Dr. Dahlquist)INCOMPLETE
- Group PowerPoint presentation (given on Tuesday, December 10, .ppt, .pptx or .pdf file) COMPLETE
- Excel spreadsheet with ANOVA results/stem formatting (.xlsx) COMPLETE
- PowerPoint of ANOVA table, screenshots of stem results (.pptx), screenshot of black and white GRNsight input network and colored GRNsight output networks COMPLETE
- Gene List and GO List files from each significant profile (.txt compressed together in a .zip file) COMPLETE
- YEASTRACT "rank by TF" results (.xlsx) COMPLETE
- GRNmap input workbook (with network adjacency matrix, .xlsx) COMPLETE
- GRNmap output workbook (.xlsx) and output plots (.jpg) zipped together COMPLETE
- MS Access database, unified by the three teams with expression tables and metadata table(s) created (.accdb) COMPLETE
- ReadMe for the database that describes the design of the database, references the sources of the data, and has a database schema diagram (.doc, .docx, .pdf) COMPLETE
- Query design for populating a GRNmap input workbook from the database (screenshot of MS Access, or SQL code, .txt) COMPLETE
- Electronic notebook corresponding to these the microarray results files (Week 12/13 and Week 15) to support reproducible research so that all manipulations of the data and files are documented so that someone else could begin with your starting file, follow the protocol, and obtain your results. INCOMPLETE
Group Report
Individual Statements of Work, Assessments, Reflections
Group Powerpoint Presentation
Sulfiknights Final Presentation
Excel Spreadsheet with ANOVA Results/STEM Formatting
Thorsen Data (Updated 12/2/19)
PowerPoint of ANOVA Table...
PowerPoint of ANOVA table and Screenshots of STEM Profile Results
Profiles 9 and 23 input network (Excel)
Profile 40 and 48 input network (Excel)
Gene List and GO List files
Zipped Gene Lists for Significant Profiles
Zipped GOLists for Significant Profiles
YEASTRACT "Rank by TF" Results
GRNmap input workbook
GRNmap Input Worksheet: Profiles 9 and 23
GRNmap Input Worksheet: Profiles 40 and 48
GRNmap output workbook and output plots
GRNmap Output Files: Profiles 9 and 23
GRNmap Output Files: Profiles 40 and 48
MS Access database
MS Access Database of Thorsen & Dahlquist data
ReadMe
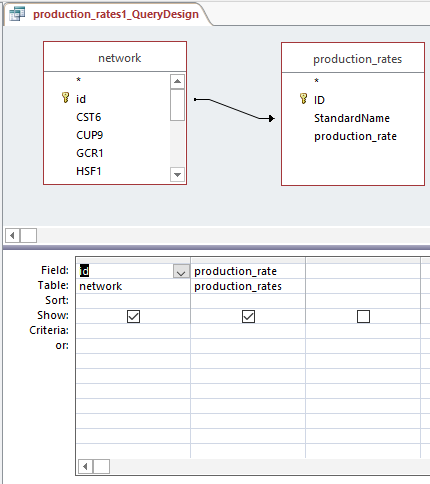
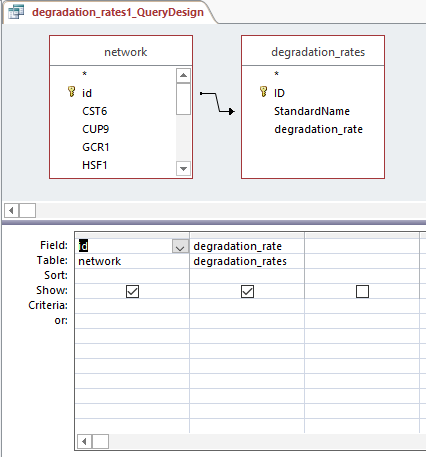
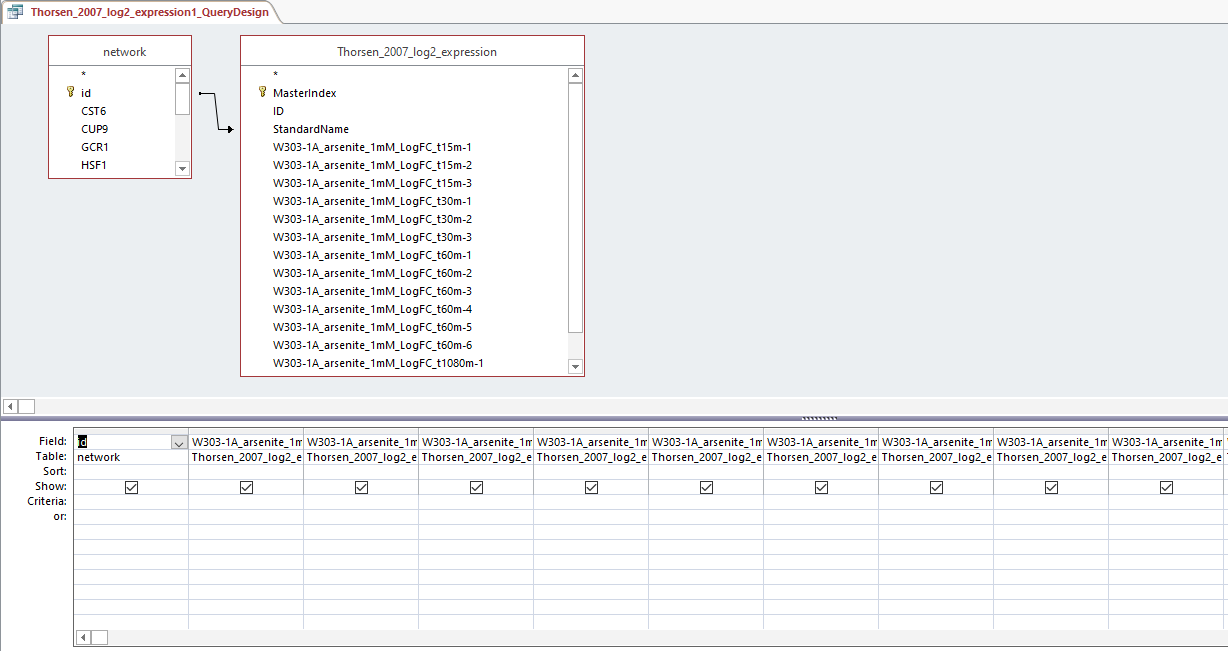
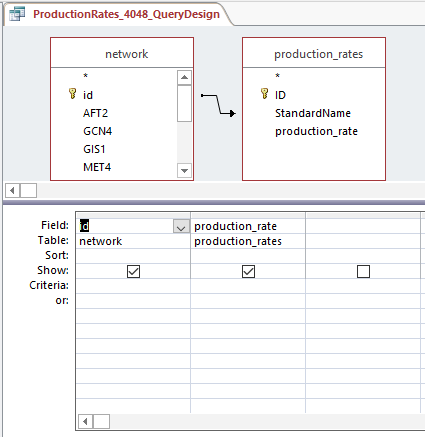
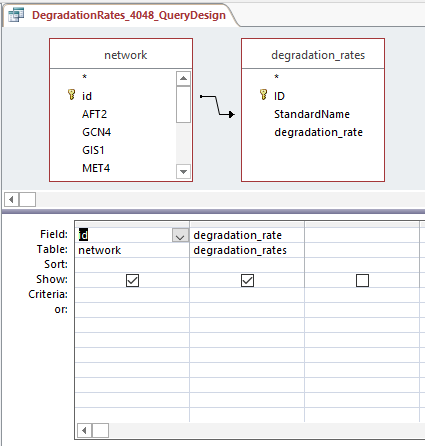
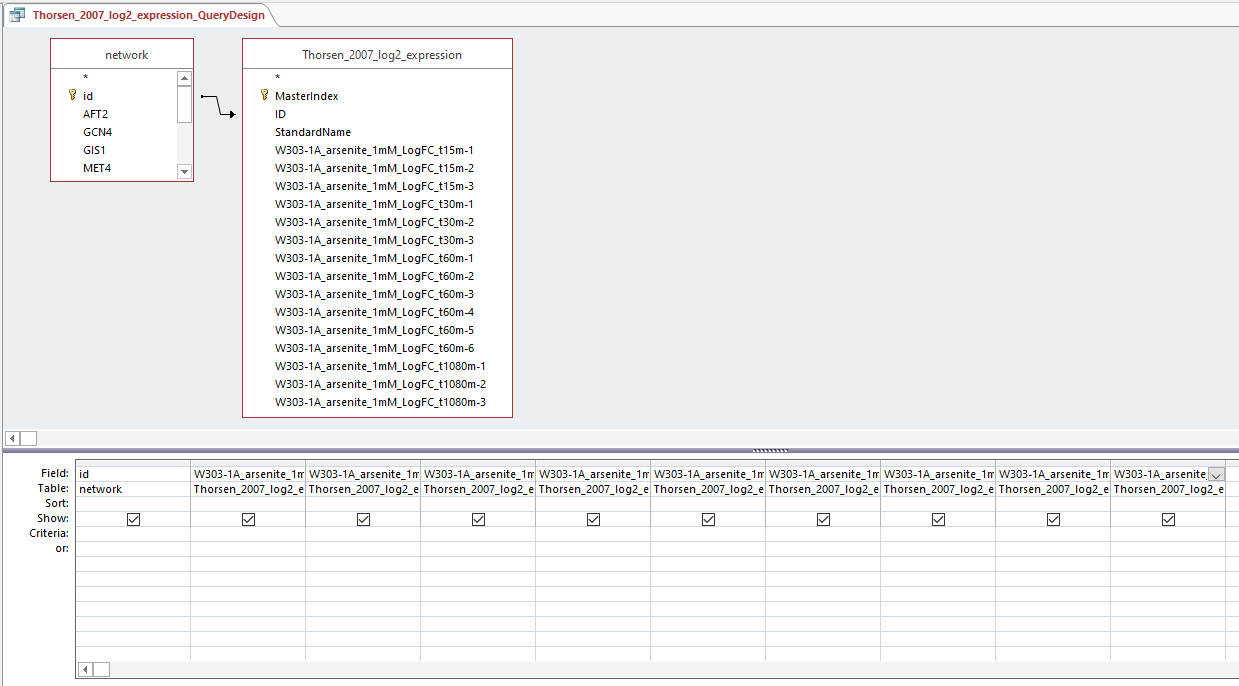
Query design for populating a GRNmap input workbook from the database
Profiles 9 and 23:
Profiles 40 and 48:
Electronic notebook corresponding to the microarray results files
Electronic notebook for Data Analysts:
Electronic notebook for Quality Assurance
Electronic notebook for Designer
Merge completed databases into a single database
BIOL_478_-_Sulfiknights_BioDB_CombinedDatabase.zip
Final Reflection
Marcus Avila's Final Reflection
Individual Assessment and Reflection
Statement of Work
- Describe exactly what you did on the project.
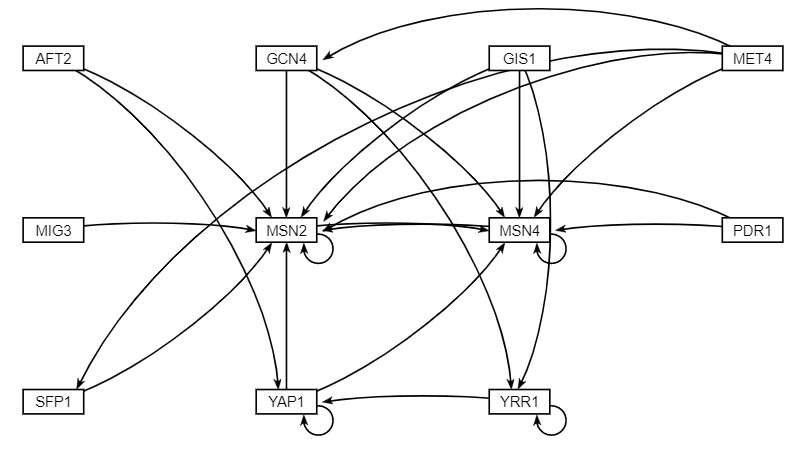
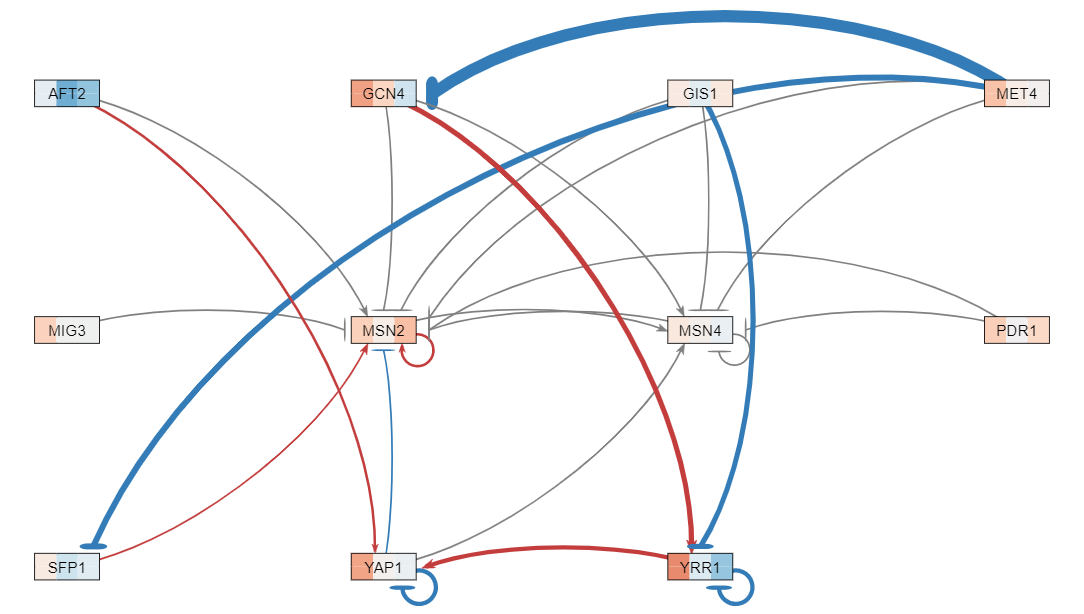
- For the project I worked with Ivy to decide how to organize the Thorsen data to run an ANOVA. After doing the ANOVA and running STEM, I then downloaded the Gene Lists for Profiles 40 & 48 and combined them to input into "Rank by TF" on YEASTRACT. Next, I selected the 15 most significant transcription factors to include on the network. After inputting the 15 transcription factors into the Generate Regulation Matrix I downloaded and transposed the Profile 40 and 48 network excel sheet. I then used GRNsight to create the weighted and unweighted network maps.
- I then created the production rate sheet, degradation rate sheet, expression data sheet, network sheet, network weights sheet, optimization parameters, and the threshold b sheet using the sample workbook provided. Finally, I input the GRNmap input workbook from the GRNmap Input Worksheet Profiles 40 and 48 into MATLAB for analysis.
Assessment of Project
- Give an objective assessment of the success of your project workflow and teamwork.
- The team did a good job making sure things were done in a timely manner and we also had very few problems meeting up outside of class time to work on the project which made the workflow much more efficient.
- What worked and what didn't work?
- Communication via text messages and scheduling meet up times worked very well. It seemed like the work load distribution was not completely equal, but this is not something we could change as a group.
- What would you do differently if you could do it all over again?
- I would ensure that each job had defined tasks or were assigned to help other members in their tasks.
- Evaluate your team’s portion of the Final Project and Group Report in the following areas:
- Content: What is the quality of the work?
- The quality of the work was good, but might have some issues because some steps were unclear and did not always parallel the steps done in the example data analysis assignment.
- Organization: Comment on the organization of the project and of your group's wiki pages.
- Our wiki page was very organized because each section included a header and files were broken up into categories/steps.
- Completeness: Did your team achieve all of the project objectives? Why or why not?
- Yes, the Microsoft Access database was successfully made after analyzing the publish microarray data.
- Content: What is the quality of the work?
Reflection on the Process
- What did you learn?
- With your head (biological or computer science principles)
- I learned about how microarray experiments can be used to study gene expression changes in response to stress.
- With your heart (personal qualities and teamwork qualities that make things work or not work)?
- I learned that meeting up is much more effective than working individually or texting when analyzing and interpreting data, as well as when working on editing presentations.
- With your hands (technical skills)?
- I learned how to input microarray data into MS Access, how to use GRNsight, YEASTRACT, and MATLAB.
- What lesson will you take away from this project that you will still use a year from now?
- I will take away the lesson that my attitude plays a major role in whether each person is equally invested in a group assignment or not.
DeLisa Madere's Final Reflection
Statement of Work
- Role as Designer
- My role in this project was a Designer. As a designer, I collaborated with the other coders to come up with a metadata sheet incorporating information regarding the citations of the paper as well as the sample-to-data sheet including the different treatments, yeast strains, timepoints, concentrations, and replicates. After creating the metadata sheet, I worked with the QAs to create the database compiling the datasets from our microarray data for the Data Analysts to analyze further. Once all the data was analyzed, I then worked with the QAs and the other coders to create a database compiling our data, the degradation rates, production rates, metadata table, as well as the other datasets from the other groups (FunGals, Skinny Genes, Dahlquist Lab) to visualize the relationships in a database schema. Each dataset was standardized under common headers such as author_publicationyear_log2_expression, and each dataset had common headers as well representing each replicate such as yeaststrain_treatment_concentration_LogFC_time-replicate. The final database is linked together through a standardized ID that each of the datasets contained.
- Links to Artifacts
Assessment of Project
Reflection on the Process
Naomi Tesfaiohannes's Final Reflection
Statement of Work
Describe exactly what you did on the project.
For this project I served as the project manager and the quality assurance guild. As project manager I communicated with Dr. Dahlquist of the steps that were made and need to continue to be made for the project. I also checked in with my team members of their progress with their roles. As QA I worked along with Joey on the datasheets to hand off to the Data Analysts. I also contributed to presenting and putting together the final presentation.
Provide references or links to artifacts of your work, such as:
Access_Database_Schema_Screenshot.PNG
Give an objective assessment of the success of your project workflow and teamwork.
What worked and what didn't work?
What worked was meeting outside of class and keeping in touch with progress by using the group chat What didn't work was sometimes not being able to answer each other's questions. Leaving us all confused.
What would you do differently if you could do it all over again?
Ask more questions about analyzing new data instead of trying to figure it out on my own.
Evaluate your team’s portion of the Final Project and Group Report in the following areas:
Content: What is the quality of the work?
Our work is a reflection of the collaborative effort from all of the team members in trying to understand the new data we reanalyzed.
Organization: Comment on the organization of the project and of your group's wiki pages.
Our group stayed organized by splitting up what sections we would work on. That way we could come together at the end of the slides to review what still needs to be understood.
Completeness: Did your team achieve all of the project objectives? Why or why not?
Yes, but at our own pace. The objectives were completed by going through the deliverables for each members.
Reflection on the Process
What did you learn?
With your head (biological or computer science principles)
Connecting the affects of arsenic to budding yeast
With your heart (personal qualities and teamwork qualities that make things work or not work)?
Teamwork and collaborating with others by contributing my own section of work with theirs to form an overall analysis of the data.
With your hands (technical skills)?
Formatting the database column titles
What lesson will you take away from this project that you will still use a year from now?
It's important to ask questions when struggling with something new, for example a database format you are not familiar with. It's not a bad thing to ask questions, and it doesn't mean you are unaware of what is happening. Questions help with understanding making it easier for the student to present on the material.