Ckaplan Week 5
Contents
- 1 Electronic Lab Notebook
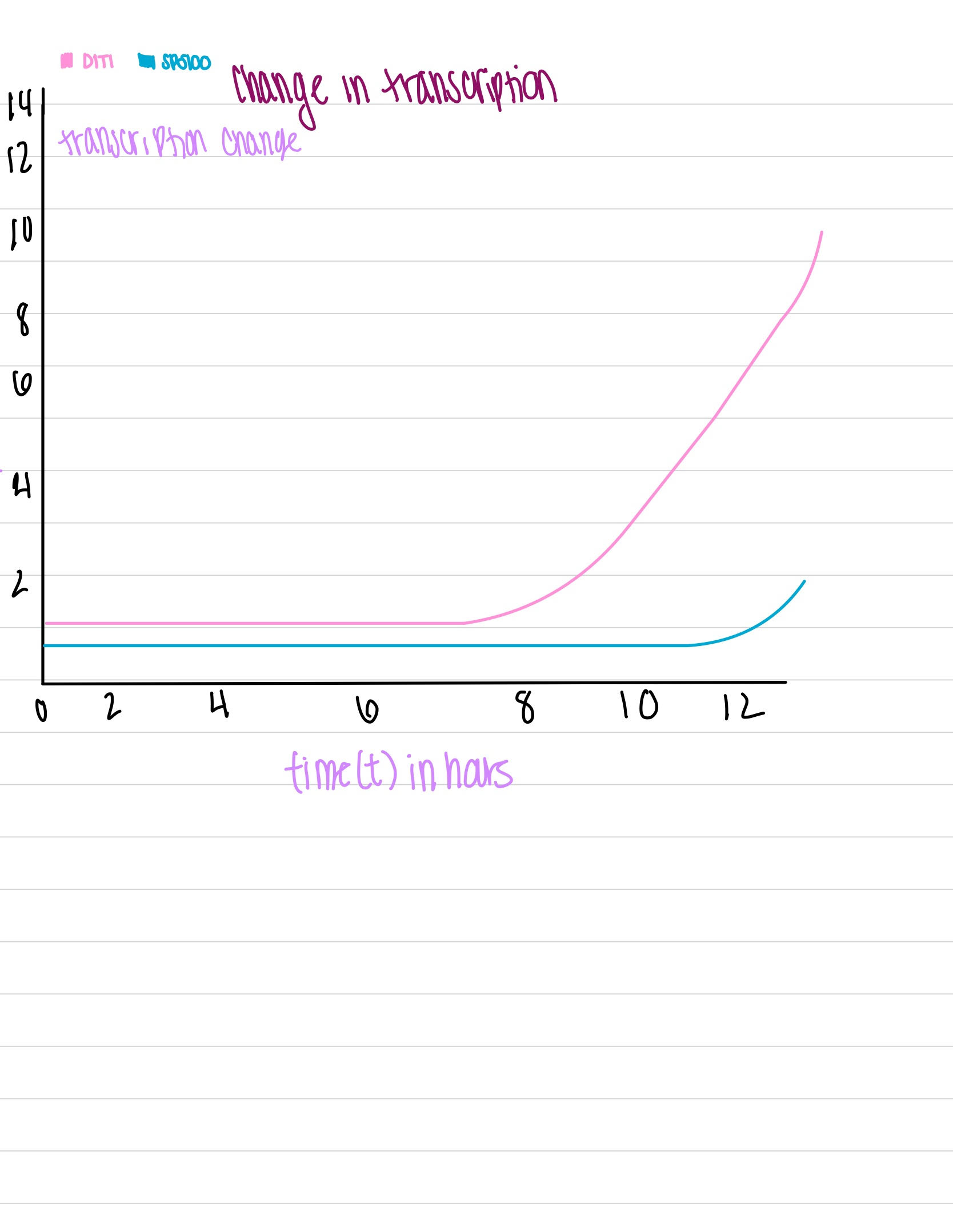
- 1.1 (Question 5, p. 110) Choose two genes from Figure 4.6b (PDF of figures on Brightspace) and draw a graph to represent the change in transcription over time. You can either create your plot in Excel and put the image up on your wiki page or you can do it by hand and upload a picture or scan.
- 1.2 (Question 6b, p. 110) Look at Figure 4.7, which depicts the loss of oxygen over time and the transcriptional response of three genes. These data are the ratios of transcription for genes X, Y, and Z during the depletion of oxygen. Using the color scale from Figure 4.6, determine the color for each ratio in Figure 4.7b. (Use the nomenclature "bright green", "medium green", "dim green", "black", "dim red", "medium red", or "bright red" for your answers.)
- 1.3 (Question 7, p. 110) Were any of the genes in Figure 4.7b transcribed similarly? If so, which ones were transcribed similarly to which ones?
- 1.4 (Question 9, p. 118) Why would most spots be yellow at the first time point? I.e., what is the technical reason that spots show up as yellow - where does the yellow color come from? And, what would be the biological reason that the experiment resulted in most spots being yellow?
- 1.5 (Question 10, p. 118) Go to the Saccharomyces Genome Database and search for the gene TEF4; you will see it is involved in translation. Look at the time point labeled OD 3.7 in Figure 4.12, and find the TEF4 spot. Over the course of this experiment, was TEF4 induced or repressed? Hypothesize why TEF4’s change in expression was part of the cell’s response to a reduction in available glucose (i.e., the only available food).
- 1.6 (Question, 11, p. 120) Why would TCA cycle genes be induced if the glucose supply is running out?
- 1.7 (Question 12, p. 120) What mechanism could the genome use to ensure genes for enzymes in a common pathway are induced or repressed simultaneously?
- 1.8 (Question 13, p. 121) Consider a microarray experiment where cells deleted for the repressor TUP1 were subjected to the same experiment of a timecourse of glucose depletion where cells at t0 (plenty of glucose available) are labeled green and cells at later timepoints (glucose depleted) are labeled red. What color would you expect the spots that represented glucose-repressed genes to be in the later time points of this experiment?
- 1.9 (Question 14, p. 121) Consider a microarray experiment where cells that overexpress the transcription factor Yap1p were subjected to the same experiment of a timecourse of glucose depletion where cells at t0 (plenty of glucose available) are labeled green and cells at later timepoints (glucose depleted) are labeled red. What color would you expect the spots that represented Yap1p target genes to be in the later time points of this experiment?
- 1.10 (Question 16, p. 121) Using the microarray data, how could you verify that you had truly deleted TUP1 or overexpressed YAP1 in the experiments described in questions 8 and 9?
- 2 Acknowledgements
- 3 References
Electronic Lab Notebook
- Purpose:
The assignment helps our class analyze and understand molecular biology experiments. It includes questions based on figures and scenarios from our course materials. Through these questions, we learn to interpret graphs, understand gene expression changes, and consider the reasons behind experimental results. Overall, it's meant to improve our understanding of molecular biology concepts and how to interpret scientific data.
Answer the following questions related to Chapter 4 of Campbell & Heyer (2003). Note that some of the questions below have been reworded from the Discovery Questions in the book:
(Question 5, p. 110) Choose two genes from Figure 4.6b (PDF of figures on Brightspace) and draw a graph to represent the change in transcription over time. You can either create your plot in Excel and put the image up on your wiki page or you can do it by hand and upload a picture or scan.
(Question 6b, p. 110) Look at Figure 4.7, which depicts the loss of oxygen over time and the transcriptional response of three genes. These data are the ratios of transcription for genes X, Y, and Z during the depletion of oxygen. Using the color scale from Figure 4.6, determine the color for each ratio in Figure 4.7b. (Use the nomenclature "bright green", "medium green", "dim green", "black", "dim red", "medium red", or "bright red" for your answers.)
X: black, dark red, black, medium green y: black, medium red, dark green, bright green z: black, dark red, dark red, dark red
(Question 7, p. 110) Were any of the genes in Figure 4.7b transcribed similarly? If so, which ones were transcribed similarly to which ones?
Genes X and Y exhibit a similar transcription pattern in Figure 4.7b where they start at the same initial point, then increase and decrease back to around the value of 1. They finally drop closer to 0 at the end.
(Question 9, p. 118) Why would most spots be yellow at the first time point? I.e., what is the technical reason that spots show up as yellow - where does the yellow color come from? And, what would be the biological reason that the experiment resulted in most spots being yellow?
In microarray experiments, the yellow color results from mixing green and red fluorescent dyes used for labeling. Initially, when glucose levels are high, most spots appear yellow because gene expression is similar under both conditions. This similarity in gene expression between conditions accounts for the yellow hue on the microarray.
(Question 10, p. 118) Go to the Saccharomyces Genome Database and search for the gene TEF4; you will see it is involved in translation. Look at the time point labeled OD 3.7 in Figure 4.12, and find the TEF4 spot. Over the course of this experiment, was TEF4 induced or repressed? Hypothesize why TEF4’s change in expression was part of the cell’s response to a reduction in available glucose (i.e., the only available food).
TEF4 was repressed over the course of the experiment.TEF4 responds to less glucose by helping cells manage energy and stress. As glucose levels decrease, TEF4 regulates how cells use energy, cope with stress, and adjust growth and division. TEF4's role helps cells adapt to low glucose levels by controlling their energy and stress responses.
(Question, 11, p. 120) Why would TCA cycle genes be induced if the glucose supply is running out?
When glucose is low, cells switch on TCA cycle genes to use other energy sources, maintain balance, and regulate metabolism, ensuring they can adapt and function properly.
(Question 12, p. 120) What mechanism could the genome use to ensure genes for enzymes in a common pathway are induced or repressed simultaneously?
The genome can use specific signals and proteins to make sure genes in a pathway are turned on or off at the same time.
(Question 13, p. 121) Consider a microarray experiment where cells deleted for the repressor TUP1 were subjected to the same experiment of a timecourse of glucose depletion where cells at t0 (plenty of glucose available) are labeled green and cells at later timepoints (glucose depleted) are labeled red. What color would you expect the spots that represented glucose-repressed genes to be in the later time points of this experiment?
In the experiment, as glucose levels decrease over time, genes that are usually repressed by glucose would likely show higher expression levels. This means that the spots representing these genes on the microarray would appear more red as time progresses.
(Question 14, p. 121) Consider a microarray experiment where cells that overexpress the transcription factor Yap1p were subjected to the same experiment of a timecourse of glucose depletion where cells at t0 (plenty of glucose available) are labeled green and cells at later timepoints (glucose depleted) are labeled red. What color would you expect the spots that represented Yap1p target genes to be in the later time points of this experiment?
During the experiment, as glucose levels decline, genes influenced by Yap1p would probably display a more intense red hue on the microarray. This indicates heightened gene activity in response to glucose depletion, as Yap1p triggers the activation of genes involved in stress response and defense against oxidative stress.
(Question 16, p. 121) Using the microarray data, how could you verify that you had truly deleted TUP1 or overexpressed YAP1 in the experiments described in questions 8 and 9?
To confirm TUP1 deletion or YAP1 overexpression with microarray data, one could compare gene expression between modified and normal samples by looking for changes in gene expression related to TUP1 or YAP1 functions. This shows genetic alterations.
Acknowledgements
I worked with my homework partner, Dean. We communicated over iMessage and helped each other with the reading questions. Dean helped me figure out questions 1-2 and I helped him with 7-10.
Except for what is noted above, this journal assignment was competed by me. Ckapla12 (talk) 21:20, 13 February 2024 (PST)
References
https://www.yeastgenome.org/locus/S000001564
Alberts et al. (2002) Molecular Biology of the Cell, Ch. 8: Microarrays Microarray animation Brown, P.O. & Botstein, D. (1999) Exploring the new world of the genome with DNA microarrays Nature Genetics 21: 33-37. https://doi.org/10.1038/4462 Campbell, A.M. and Heyer, L.J. (2003), “Chapter 4: Basic Research with DNA Microarrays”, in Discovering Genomics, Proteomics, and Bioinformatics, Cold Spring Harbor Laboratory Press, pp. 107-124. (Available on Brightspace) DeRisi, J.L., Iyer, V.R., and Brown, P.O. (1997) Exploring the Metabolic and Genetic Control of Gene Expression on a Genomic Scale. Science 278: 680-686. DOI: https://doi.org/10.1126/science.278.5338.680