SIR2 Week 3
Contents
- 1 Summary:
- 1.1 What is the standard name, systematic name, and name description for your gene (from SGD)?
- 1.2 What is the gene ID (identifier) for your gene in all four databases (SGD, NCBI Gene, Ensembl, UniProt)?
- 1.3 Provide hyperlinks to the specific pages for your gene in each of the above databases.
- 1.4 What is the DNA sequence of your gene?
- 1.5 What is the protein sequence corresponding to your gene?
- 1.6 Go to the ExPASy tool and translate the DNA sequence of your gene. Which reading frame encodes the protein sequence? Take a screenshot of your results, display it on your wiki page, and state which frame it is.
- 1.7 What is the function of your gene?
- 1.8 What was different about the information provided about your gene in each of the parent databases?
- 1.9 Were there differences in content, the information or data itself?
- 1.10 Were there differences in presentation of the information?
- 1.11 Why did you choose your particular gene? i.e., why is it interesting to you and your partner? Include an image related to your gene (be careful that you do not violate any copyright restrictions!)
- 1.12 Acknowledgments and References
Summary:
The gene Sir 2 is part of the Sirtuin family. The Sirtuin family plays a role in lifespan/healthspan according to an article called "Sirtuins, a promising target in slowing down the ageing process."
Sir2 is a critical part of cellular function. It has a role in important functions such as regulating lifespan and contributing to "genome stability". It also deacetylates nuclear proteins, which means it causes the removal of an acetyl group.
According to the summary in the NCBI Database it:
"Enables NAD-dependent histone deacetylase activity (H3-K14 specific) and NAD-dependent histone deacetylase activity (H4-K16 specific). Contributes to nucleosome binding activity. Involved in several processes, including establishment of protein-containing complex localization to telomere; negative regulation of macromolecule metabolic process; and regulation of DNA stability. Located in chromosome, telomeric region; heterochromatin; and nucleolus. Part of RENT complex. Used to study Alzheimer's disease; Parkinson's disease; diabetes mellitus; and prostate cancer. Human ortholog(s) of this gene implicated in Huntington's disease. Orthologous to human SIRT1 (sirtuin 1)."
Research indicates that the activity of sirtuin proteins, including Sir2/SIRT1, plays a role in modulating lifespan. One of the primary mechanisms involves the regulation of cellular stress responses and DNA repair processes. Sirtuins are involved in maintaining genomic stability and promoting efficient DNA repair, which are crucial for preventing the accumulation of DNA damage over time.
Sir2 also has been found to regulate key metabolic pathways including glucose, lipid metabolism and mitochondrial function. By regulating these pathways, Sir2 can influence energy production, oxidative stress levels, and overall metabolic health, which impacts lifespan.
NAD-dependent histone deacetylase involved in chromatin assembly, organization and silencing. Also involved in sister chromatid cohesion, double-strand break repair, and the tethering of telomeres at the nuclear periphery (SGD).
Conserved NAD+ dependent histone deacetylase of the Sirtuin family; deacetylation targets are primarily nuclear proteins; required for telomere hypercluster formation in quiescent yeast cells; involved in regulation of lifespan; plays roles in silencing at HML, HMR, telomeres, and rDNA; negatively regulates initiation of DNA replication; functions as regulator of autophagy like mammalian homolog SIRT1, and also of mitophagy [Source:SGD;Acc:S000002200]
What is the standard name, systematic name, and name description for your gene (from SGD)?
-Standard Name: SIR2 1
-Systematic Name: YDL042C
Description from SGD: Conserved NAD+ dependent histone deacetylase of the Sirtuin family; deacetylation targets are primarily nuclear proteins; required for telomere hypercluster formation in quiescent yeast cells; involved in regulation of lifespan; plays roles in silencing at HML, HMR, telomeres, and rDNA; negatively regulates initiation of DNA replication, suppressing early firing of multicopy rDNA origins; functions as regulator of autophagy like mammalian homolog SIRT1, and also of mitophagy.
What is the gene ID (identifier) for your gene in all four databases (SGD, NCBI Gene, Ensembl, UniProt)?
SGD: S000002200
Uniprot: P06700
Ensembl: YDL042C
NCBI: 851520
Provide hyperlinks to the specific pages for your gene in each of the above databases.
https://www.yeastgenome.org/locus/S000002200
https://www.uniprot.org/uniprotkb/P06700/entry
https://www.ncbi.nlm.nih.gov/gene/851520
What is the DNA sequence of your gene?
1 ATGACCATCC CACATATGAA ATACGCCGTA TCAAAGACTA GCGAAAATAA GGTTTCAAAT 61 ACAGTAAGCC CCACACAAGA TAAAGACGCG ATCAGAAAAC AACCCGATGA CATTATAAAT 121 AATGATGAAC CTTCACATAA GAAGATAAAA GTAGCACAGC CGGATTCCTT GAGGGAAACC 181 AACACAACAG ATCCACTTGG GCACACTAAA GCTGCGCTCG GAGAAGTGGC ATCGATGGAG 241 CTCAAACCAA CTAATGACAT GGATCCCTTG GCAGTGTCAG CAGCTTCAGT AGTGTCAATG 301 TCCAATGACG TTTTGAAACC AGAGACGCCC AAGGGGCCAA TCATAATCAG TAAAAACCCA 361 TCAAATGGTA TTTTCTATGG TCCCTCCTTC ACTAAACGAG AGTCTCTCAA TGCTCGAATG 421 TTTCTGAAAT ACTATGGTGC ACACAAATTT TTAGACACTT ACCTCCCCGA GGATTTGAAC 481 TCGTTATACA TTTACTATCT TATCAAGTTG CTAGGCTTTG AAGTTAAAGA TCAAGCGCTT 541 ATCGGCACCA TCAACAGTAT TGTCCATATC AACTCGCAAG AGCGTGTTCA AGATTTGGGA 601 AGTGCAATAT CTGTCACAAA TGTTGAAGAC CCATTGGCAA AAAAGCAAAC AGTTCGTCTA 661 ATCAAAGATT TGCAAAGAGC AATTAACAAA GTTCTATGTA CAAGATTAAG ATTATCCAAT 721 TTTTTCACTA TTGATCATTT TATTCAAAAA TTACATACCG CTAGAAAAAT TTTGGTCCTG 781 ACTGGTGCAG GTGTTTCAAC TTCATTAGGG ATCCCGGACT TCAGATCTTC TGAGGGGTTC 841 TATTCAAAGA TCAAACATTT GGGGCTCGAT GATCCCCAAG ACGTTTTCAA TTACAATATA 901 TTTATGCACG ACCCCTCTGT TTTCTATAAT ATTGCCAATA TGGTTTTACC TCCAGAAAAA 961 ATTTATTCTC CATTGCATAG TTTCATTAAG ATGCTACAAA TGAAAGGGAA ATTATTGAGA 1021 AATTATACTC AAAACATTGA TAATTTGGAA TCTTATGCGG GAATAAGCAC AGATAAACTG 1081 GTGCAGTGCC ATGGCTCTTT TGCTACTGCC ACCTGCGTTA CCTGCCATTG GAACCTACCC 1141 GGTGAGAGGA TATTTAATAA AATTAGAAAC CTCGAACTTC CACTATGCCC GTACTGTTAC 1201 AAAAAAAGAA GAGAATATTT CCCAGAGGGA TATAATAATA AAGTAGGTGT TGCTGCATCA 1261 CAGGGTTCAA TGTCGGAAAG GCCTCCATAT ATCCTTAACT CATATGGCGT TCTCAAACCA 1321 GATATCACAT TCTTTGGCGA AGCACTGCCA AATAAATTTC ATAAGAGCAT TCGCGAAGAT 1381 ATCTTAGAAT GTGATTTGTT GATTTGCATT GGGACAAGTT TAAAAGTAGC GCCAGTGTCT 1441 GAAATCGTAA ACATGGTTCC TTCCCACGTT CCCCAAGTCC TGATTAATCG TGATCCCGTC 1501 AAGCACGCAG AATTTGATTT ATCTCTTTTG GGGTACTGTG ATGACATTGC AGCTATGGTA 1561 GCCCAAAAAT GTGGCTGGAC GATTCCGCAT AAGAAATGGA ACGATTTGAA GAACAAGAAC 1621 TTTAAATGCC AAGAGAAGGA TAAGGGCGTG TATGTCGTTA CATCAGATGA ACATCCCAAA 1681 ACCCTCTAA
What is the protein sequence corresponding to your gene?
1 MTIPHMKYAV SKTSENKVSN TVSPTQDKDA IRKQPDDIIN NDEPSHKKIK VAQPDSLRET 61 NTTDPLGHTK AALGEVASME LKPTNDMDPL AVSAASVVSM SNDVLKPETP KGPIIISKNP 121 SNGIFYGPSF TKRESLNARM FLKYYGAHKF LDTYLPEDLN SLYIYYLIKL LGFEVKDQAL 181 IGTINSIVHI NSQERVQDLG SAISVTNVED PLAKKQTVRL IKDLQRAINK VLCTRLRLSN 241 FFTIDHFIQK LHTARKILVL TGAGVSTSLG IPDFRSSEGF YSKIKHLGLD DPQDVFNYNI 301 FMHDPSVFYN IANMVLPPEK IYSPLHSFIK MLQMKGKLLR NYTQNIDNLE SYAGISTDKL 361 VQCHGSFATA TCVTCHWNLP GERIFNKIRN LELPLCPYCY KKRREYFPEG YNNKVGVAAS 421 QGSMSERPPY ILNSYGVLKP DITFFGEALP NKFHKSIRED ILECDLLICI GTSLKVAPVS 481 EIVNMVPSHV PQVLINRDPV KHAEFDLSLL GYCDDIAAMV AQKCGWTIPH KKWNDLKNKN 541 FKCQEKDKGV YVVTSDEHPK TL*
Go to the ExPASy tool and translate the DNA sequence of your gene. Which reading frame encodes the protein sequence? Take a screenshot of your results, display it on your wiki page, and state which frame it is.
Reading frame 1 codes for the protein sequence.
This Open reading frame specifically:
MTIPHMKYAVSKTSENKVSNTVSPTQDKDAIRKQPDDIINNDEPSHKKIKVAQPDSLRETNTTDPLGHTKAALGEVASMELKPTNDMDPLAVSAASVVSMSNDVLKPETPKGPIIISKNPSNGIFYGPSFTKRESLNARMFLKYYGAHKFL DTYLPEDLNSLYIYYLIKLLGFEVKDQALIGTINSIVHINSQERVQDLGSAISVTNVEDPLAKKQTVRLIKDLQRAINKVLCTRLRLSNFFTIDHFIQKLHTARKILVLTGAGVSTSLGIPDFRSSEGFYSKIKHLGLDDPQDVFNYNIFM HDPSVFYNIANMVLPPEKIYSPLHSFIKMLQMKGKLLRNYTQNIDNLESYAGISTDKLVQCHGSFATATCVTCHWNLPGERIFNKIRNLELPLCPYCYKKRREYFPEGYNNKVGVAASQGSMSERPPYILNSYGVLKPDITFFGEALPNKF HKSIREDILECDLLICIGTSLKVAPVSEIVNMVPSHVPQVLINRDPVKHAEFDLSLLGYCDDIAAMVAQKCGWTIPHKKWNDLKNKNFKCQEKDKGVYVVTSDEHPKTL
What is the function of your gene?
Sir2 acts as a NAD+-dependent histone deacetylase. It is involved in gene silencing at specific chromosomal sites like telomeres and loci, and in preserving genomic stability through DNA repair mechanisms. Sir2 influences the regulation of lifespan with increased expression or activity often associated with lifespan extension. It also modulates metabolic pathways such as glucose and lipid metabolism, as well as mitochondrial function, impacting energy generation and cellular health.
What was different about the information provided about your gene in each of the parent databases?
In one database it included not only the Exon, but the surrounding DNA, Intron. (Ensemble) But the database Charlotte worked with only had the Exon.
Were there differences in content, the information or data itself?
There was a lot of data. There was differences in the total amount of content, but the data did not seem to differ. I don't know where to look other than the Gene in terms of data.
Were there differences in presentation of the information?
There were differences in presentation and organization of the data. Ensembl was the most straightforward and helpful. It had a nice side bar. NCBI was thorough and organized, but it was not intuitive and searching through took a lot of trial and error. I really like that NCBI has so many articles for free to access but I do with the navigation was easier. UniProt was also very intuitive and well organized. I really like the side navigation bar that scrolls through as you scroll. It also had a sleek presentation and felt on par with Ensembl. YeastGenome was also great. Really enjoyed the fact that there were lots of resources and articles related to the topic provided directly on the page. They had history with references and that was something Ensemble did not have.
We chose our gene because we wanted to learn about a gene that fit two qualifications:
- We decided our gene had to have a related human gene.
- We wanted our gene to play a role in aging and if we could also have a gene related to disease that was a plus.
This gene satisfied both of those criteria directly.
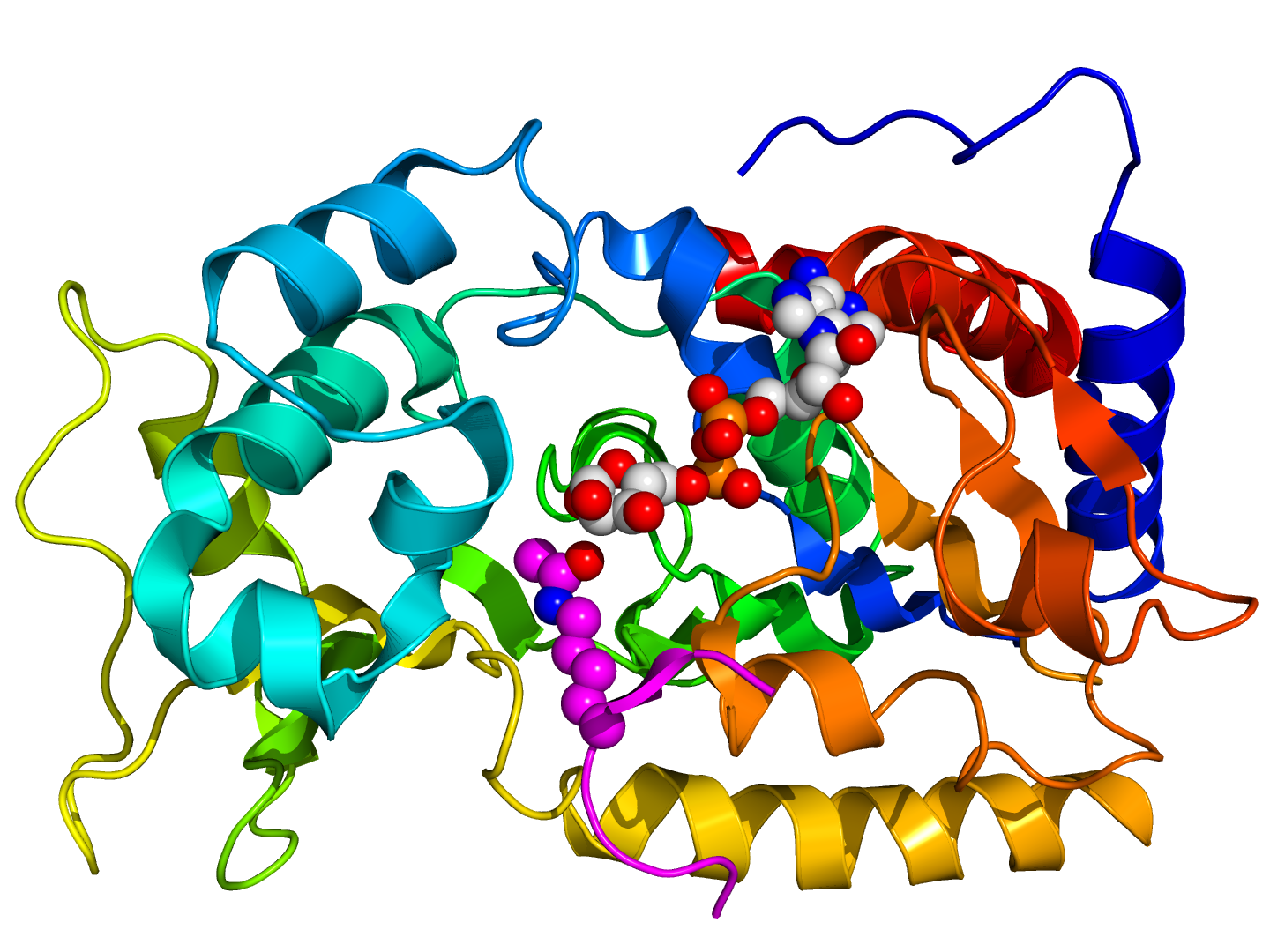
Please note the 3D structure of SIR2 is shown on the structure section of it's UniProt entry. The issue is that the Alphafold rendering that is shown is labeled as low confidence, and the x-ray determined structures available from PDB do not show the entire section from positions 1-562, they only show chunks of the positions.
Due to this, we included an image of SIR2 from the Sirtuin Wikipedia page.
Acknowledgments and References
Guarente, L. (2013). Calorie restriction and sirtuins revisited. Genes & Development, 27(19), 2072–2085. https://doi.org/10.1101/gad.227439.113 Kennedy, B. K., & Lamming, D. W. (2016). The Mechanistic Target of Rapamycin: The Grand ConducTOR of Metabolism and Aging. Cell Metabolism, 23(6), 990–1003. https://doi.org/10.1016/j.cmet.2016.05.009 Bonkowski, M. S., & Sinclair, D. A. (2016). Slowing ageing by design: the rise of NAD+ and sirtuin-activating compounds. Nature Reviews Molecular Cell Biology, 17(11), 679–690. https://doi.org/10.1038/nrm.2016.93
Sebastián, C., Satterstrom, F. K., Haigis, M. C., & Mostoslavsky, R. (2017). From sirtuin biology to human diseases: An update. Journal of Biological Chemistry, 292(53), 22352–22360. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5514220/
https://www.yeastgenome.org/locus/S000002200
https://www.uniprot.org/taxonomy/559292
https://www.ncbi.nlm.nih.gov/gene/851520
https://en.wikipedia.org/wiki/Sirtuin
I want to provide an acknowledgement to the Wikipedia user Boghog, this user created the image I took from wikipedia of the structure of SIR 2. Link is here: https://commons.wikimedia.org/wiki/User:Boghog
LMU BioDb 2024. (2024). Week 3. Retrieved January 31, 2024, from https://xmlpipedb.cs.lmu.edu/biodb/spring2024/index.php/Week_3
Except for what is noted above, this individual journal entry was completed by me and not copied from another source. Ckapla12 (talk) 16:13, 31 January 2024 (PST)
Except for what is noted above or was the part completed by Charlotte, this journal entry was completed by me and not copied from another source. Asandle1 (talk) 22:45, 31 January 2024 (PST)

